Collaborative Filtering#
pimmslearn - INFO Experiment 03 - Analysis of latent spaces and performance comparisions
Papermill script parameters:
# files and folders
# Datasplit folder with data for experiment
folder_experiment: str = 'runs/example'
folder_data: str = '' # specify data directory if needed
file_format: str = 'csv' # change default to pickled files
# training
epochs_max: int = 20 # Maximum number of epochs
# early_stopping:bool = True # Wheather to use early stopping or not
patience: int = 1 # Patience for early stopping
batch_size: int = 32_768 # Batch size for training (and evaluation)
cuda: bool = True # Use the GPU for training?
# model
# Dimensionality of encoding dimension (latent space of model)
latent_dim: int = 10
sample_idx_position: int = 0 # position of index which is sample ID
model: str = 'CF' # model name
model_key: str = 'CF' # potentially alternative key for model (grid search)
save_pred_real_na: bool = True # Save all predictions for missing values
# Parameters
model = "CF"
latent_dim = 50
batch_size = 1024
epochs_max = 100
sample_idx_position = 0
cuda = False
save_pred_real_na = True
fn_rawfile_metadata = "https://raw.githubusercontent.com/RasmussenLab/njab/HEAD/docs/tutorial/data/alzheimer/meta.csv"
folder_experiment = "runs/alzheimer_study"
model_key = "CF"
Some argument transformations
{'folder_experiment': 'runs/alzheimer_study',
'folder_data': '',
'file_format': 'csv',
'epochs_max': 100,
'patience': 1,
'batch_size': 1024,
'cuda': False,
'latent_dim': 50,
'sample_idx_position': 0,
'model': 'CF',
'model_key': 'CF',
'save_pred_real_na': True,
'fn_rawfile_metadata': 'https://raw.githubusercontent.com/RasmussenLab/njab/HEAD/docs/tutorial/data/alzheimer/meta.csv'}
{'batch_size': 1024,
'cuda': False,
'data': Path('runs/alzheimer_study/data'),
'epochs_max': 100,
'file_format': 'csv',
'fn_rawfile_metadata': 'https://raw.githubusercontent.com/RasmussenLab/njab/HEAD/docs/tutorial/data/alzheimer/meta.csv',
'folder_data': '',
'folder_experiment': Path('runs/alzheimer_study'),
'latent_dim': 50,
'model': 'CF',
'model_key': 'CF',
'out_figures': Path('runs/alzheimer_study/figures'),
'out_folder': Path('runs/alzheimer_study'),
'out_metrics': Path('runs/alzheimer_study'),
'out_models': Path('runs/alzheimer_study'),
'out_preds': Path('runs/alzheimer_study/preds'),
'patience': 1,
'sample_idx_position': 0,
'save_pred_real_na': True}
Some naming conventions
Load data in long format#
pimmslearn.io.datasplits - INFO Loaded 'train_X' from file: runs/alzheimer_study/data/train_X.csv
pimmslearn.io.datasplits - INFO Loaded 'val_y' from file: runs/alzheimer_study/data/val_y.csv
pimmslearn.io.datasplits - INFO Loaded 'test_y' from file: runs/alzheimer_study/data/test_y.csv
data is loaded in long format
Sample ID protein groups
Sample_000 A0A024QZX5;A0A087X1N8;P35237 15.912
A0A024R0T9;K7ER74;P02655 16.852
A0A024R3W6;A0A024R412;O60462;O60462-2;O60462-3;O60462-4;O60462-5;Q7LBX6;X5D2Q8 15.570
A0A024R644;A0A0A0MRU5;A0A1B0GWI2;O75503 16.481
A0A075B6H7 17.301
...
Sample_209 Q9Y6R7 19.275
Q9Y6X5 15.732
Q9Y6Y8;Q9Y6Y8-2 19.577
Q9Y6Y9 11.042
S4R3U6 11.791
Name: intensity, Length: 226809, dtype: float64
Infer index names from long format
pimmslearn - INFO sample_id = 'Sample ID', single feature: index_column = 'protein groups'
Use some simulated missing for evaluation#
The validation simulated NA is used to by all models to evaluate training performance.
| observed | ||
|---|---|---|
| Sample ID | protein groups | |
| Sample_158 | Q9UN70;Q9UN70-2 | 14.630 |
| Sample_050 | Q9Y287 | 15.755 |
| Sample_107 | Q8N475;Q8N475-2 | 15.029 |
| Sample_199 | P06307 | 19.376 |
| Sample_067 | Q5VUB5 | 15.309 |
| ... | ... | ... |
| Sample_111 | F6SYF8;Q9UBP4 | 22.822 |
| Sample_002 | A0A0A0MT36 | 18.165 |
| Sample_049 | Q8WY21;Q8WY21-2;Q8WY21-3;Q8WY21-4 | 15.525 |
| Sample_182 | Q8NFT8 | 14.379 |
| Sample_123 | Q16853;Q16853-2 | 14.504 |
12600 rows × 1 columns
| observed | |
|---|---|
| count | 12,600.000 |
| mean | 16.339 |
| std | 2.741 |
| min | 7.209 |
| 25% | 14.412 |
| 50% | 15.935 |
| 75% | 17.910 |
| max | 30.140 |
Collaborative Filtering#
save custom collab batch size (increase AE batch size by a factor), could be setup separately.
the test data is used to evaluate the performance after training
Args:
{'n_factors': 50, 'y_range': (7, 31)}
Training#
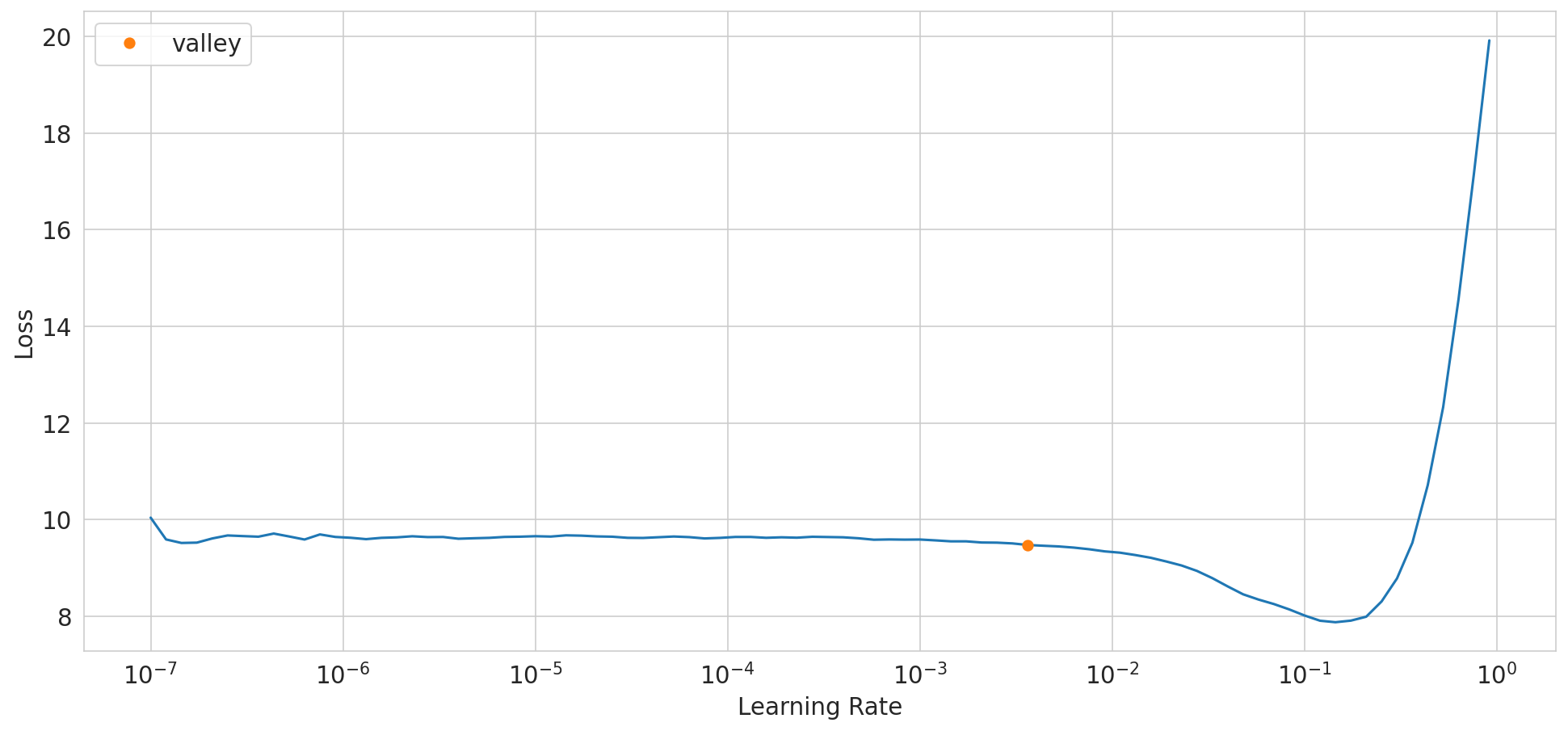
suggested_lr.valley = 0.00363
No improvement since epoch 15: early stopping
pimmslearn.plotting - INFO Saved Figures to runs/alzheimer_study/figures/collab_training
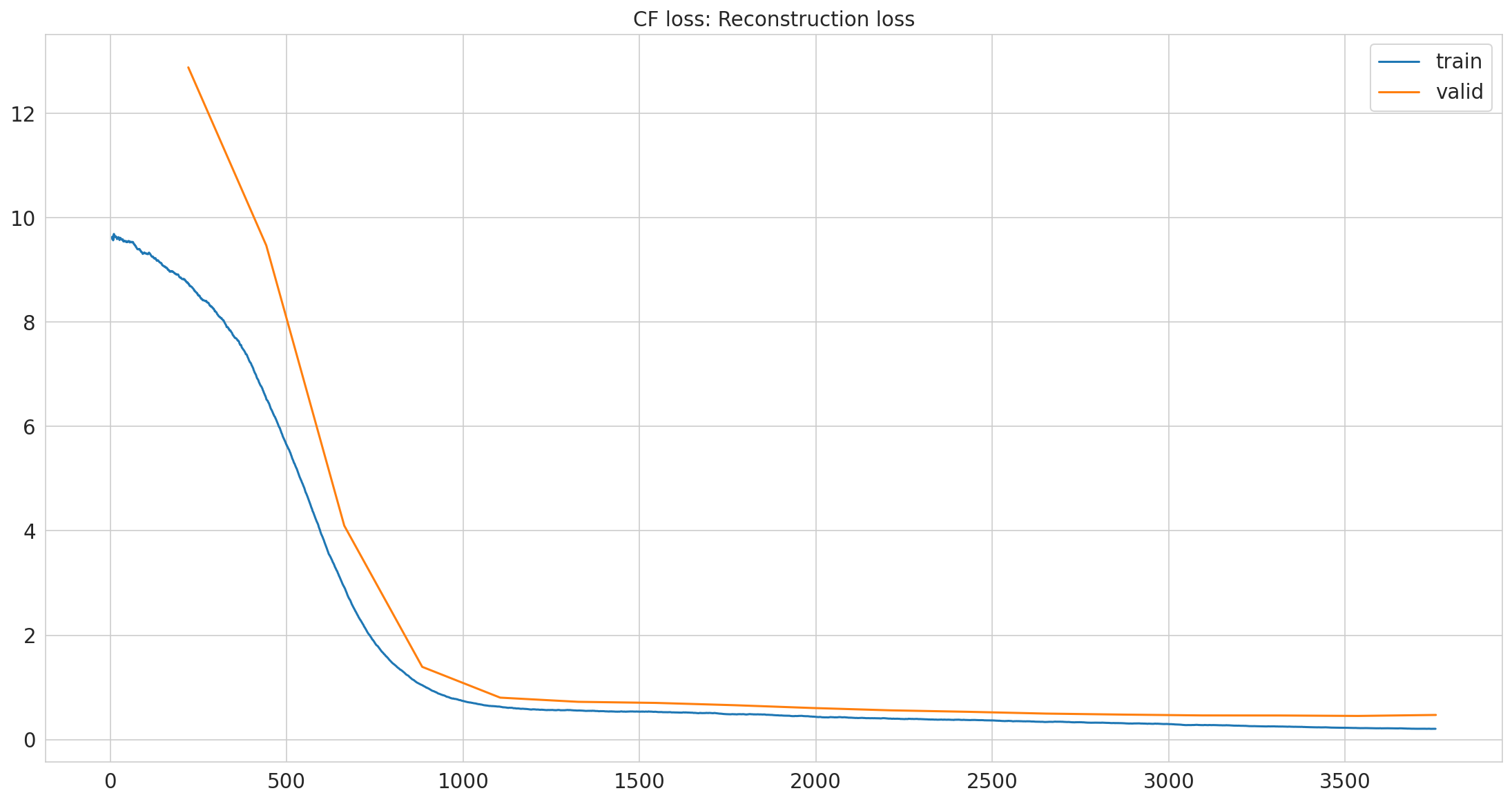
| epoch | train_loss | valid_loss | time |
|---|---|---|---|
| 0 | 8.750916 | 12.880796 | 00:01 |
| 1 | 6.556331 | 9.468315 | 00:01 |
| 2 | 2.935405 | 4.100215 | 00:01 |
| 3 | 1.047107 | 1.393667 | 00:01 |
| 4 | 0.630373 | 0.803804 | 00:01 |
| 5 | 0.556590 | 0.723707 | 00:01 |
| 6 | 0.532088 | 0.702849 | 00:01 |
| 7 | 0.486000 | 0.659230 | 00:01 |
| 8 | 0.441405 | 0.605385 | 00:01 |
| 9 | 0.401426 | 0.560744 | 00:01 |
| 10 | 0.374354 | 0.532496 | 00:01 |
| 11 | 0.337384 | 0.498261 | 00:01 |
| 12 | 0.313225 | 0.478834 | 00:01 |
| 13 | 0.277879 | 0.463119 | 00:01 |
| 14 | 0.250062 | 0.461858 | 00:01 |
| 15 | 0.220919 | 0.453349 | 00:01 |
| 16 | 0.206850 | 0.472183 | 00:01 |
Predictions#
Compare simulated_na data predictions to original values
| observed | CF | ||
|---|---|---|---|
| Sample ID | protein groups | ||
| Sample_158 | Q9UN70;Q9UN70-2 | 14.630 | 15.653 |
| Sample_050 | Q9Y287 | 15.755 | 16.477 |
| Sample_107 | Q8N475;Q8N475-2 | 15.029 | 14.060 |
| Sample_199 | P06307 | 19.376 | 19.088 |
| Sample_067 | Q5VUB5 | 15.309 | 14.916 |
| ... | ... | ... | ... |
| Sample_111 | F6SYF8;Q9UBP4 | 22.822 | 22.951 |
| Sample_002 | A0A0A0MT36 | 18.165 | 16.263 |
| Sample_049 | Q8WY21;Q8WY21-2;Q8WY21-3;Q8WY21-4 | 15.525 | 15.158 |
| Sample_182 | Q8NFT8 | 14.379 | 14.092 |
| Sample_123 | Q16853;Q16853-2 | 14.504 | 14.647 |
12600 rows × 2 columns
select test data predictions
| observed | CF | ||
|---|---|---|---|
| Sample ID | protein groups | ||
| Sample_000 | A0A075B6P5;P01615 | 17.016 | 17.612 |
| A0A087X089;Q16627;Q16627-2 | 18.280 | 17.963 | |
| A0A0B4J2B5;S4R460 | 21.735 | 22.574 | |
| A0A140T971;O95865;Q5SRR8;Q5SSV3 | 14.603 | 15.219 | |
| A0A140TA33;A0A140TA41;A0A140TA52;P22105;P22105-3;P22105-4 | 16.143 | 16.540 | |
| ... | ... | ... | ... |
| Sample_209 | Q96ID5 | 16.074 | 16.190 |
| Q9H492;Q9H492-2 | 13.173 | 13.659 | |
| Q9HC57 | 14.207 | 14.216 | |
| Q9NPH3;Q9NPH3-2;Q9NPH3-5 | 14.962 | 15.142 | |
| Q9UGM5;Q9UGM5-2 | 16.871 | 16.523 |
12600 rows × 2 columns
Data in wide format#
Autoencoder need data in wide format
| protein groups | A0A024QZX5;A0A087X1N8;P35237 | A0A024R0T9;K7ER74;P02655 | A0A024R3W6;A0A024R412;O60462;O60462-2;O60462-3;O60462-4;O60462-5;Q7LBX6;X5D2Q8 | A0A024R644;A0A0A0MRU5;A0A1B0GWI2;O75503 | A0A075B6H7 | A0A075B6H9 | A0A075B6I0 | A0A075B6I1 | A0A075B6I6 | A0A075B6I9 | ... | Q9Y653;Q9Y653-2;Q9Y653-3 | Q9Y696 | Q9Y6C2 | Q9Y6N6 | Q9Y6N7;Q9Y6N7-2;Q9Y6N7-4 | Q9Y6R7 | Q9Y6X5 | Q9Y6Y8;Q9Y6Y8-2 | Q9Y6Y9 | S4R3U6 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sample ID | |||||||||||||||||||||
| Sample_000 | 15.912 | 16.852 | 15.570 | 16.481 | 17.301 | 20.246 | 16.764 | 17.584 | 16.988 | 20.054 | ... | 16.012 | 15.178 | NaN | 15.050 | 16.842 | NaN | NaN | 19.563 | NaN | 12.805 |
| Sample_001 | NaN | 16.874 | 15.519 | 16.387 | NaN | 19.941 | 18.786 | 17.144 | NaN | 19.067 | ... | 15.528 | 15.576 | NaN | 14.833 | 16.597 | 20.299 | 15.556 | 19.386 | 13.970 | 12.442 |
| Sample_002 | 16.111 | NaN | 15.935 | 16.416 | 18.175 | 19.251 | 16.832 | 15.671 | 17.012 | 18.569 | ... | 15.229 | 14.728 | 13.757 | 15.118 | 17.440 | 19.598 | 15.735 | 20.447 | 12.636 | 12.505 |
| Sample_003 | 16.107 | 17.032 | 15.802 | 16.979 | 15.963 | 19.628 | 17.852 | 18.877 | 14.182 | 18.985 | ... | 15.495 | 14.590 | 14.682 | 15.140 | 17.356 | 19.429 | NaN | 20.216 | NaN | 12.445 |
| Sample_004 | 15.603 | 15.331 | 15.375 | 16.679 | NaN | 20.450 | 18.682 | 17.081 | 14.140 | 19.686 | ... | 14.757 | NaN | NaN | 15.256 | 17.075 | 19.582 | 15.328 | NaN | 13.145 | NaN |
5 rows × 1421 columns
Validation data#
all measured (identified, observed) peptides in validation data
Does not make to much sense to compare collab and AEs, as the setup differs of training and validation data differs
The simulated NA for the validation step are real test data (not used for training nor early stopping)
added_metrics = d_metrics.add_metrics(val_pred_simulated_na, 'valid_simulated_na')
added_metrics
Selected as truth to compare to: observed
{'CF': {'MSE': 0.4721825960916124,
'MAE': 0.464989392779898,
'N': 12600,
'prop': 1.0}}
Test Datasplit#
Simulated NAs : Artificially created NAs. Some data was sampled and set explicitly to misssing before it was fed to the model for reconstruction.
added_metrics = d_metrics.add_metrics(test_pred_simulated_na, 'test_simulated_na')
added_metrics
Selected as truth to compare to: observed
{'CF': {'MSE': 0.4933668310969956,
'MAE': 0.46818207034951376,
'N': 12600,
'prop': 1.0}}
Save all metrics as json
pimmslearn.io.dump_json(d_metrics.metrics, args.out_metrics /
f'metrics_{args.model_key}.json')
metrics_df = models.get_df_from_nested_dict(
d_metrics.metrics, column_levels=['model', 'metric_name']).T
metrics_df
| subset | valid_simulated_na | test_simulated_na | |
|---|---|---|---|
| model | metric_name | ||
| CF | MSE | 0.472 | 0.493 |
| MAE | 0.465 | 0.468 | |
| N | 12,600.000 | 12,600.000 | |
| prop | 1.000 | 1.000 |
Save predictions#
Config#
{'M': 1421,
'batch_size': 1024,
'cuda': False,
'data': Path('runs/alzheimer_study/data'),
'epoch_trained': 17,
'epochs_max': 100,
'file_format': 'csv',
'fn_rawfile_metadata': 'https://raw.githubusercontent.com/RasmussenLab/njab/HEAD/docs/tutorial/data/alzheimer/meta.csv',
'folder_data': '',
'folder_experiment': Path('runs/alzheimer_study'),
'latent_dim': 50,
'model': 'CF',
'model_key': 'CF',
'n_params': 83283,
'out_figures': Path('runs/alzheimer_study/figures'),
'out_folder': Path('runs/alzheimer_study'),
'out_metrics': Path('runs/alzheimer_study'),
'out_models': Path('runs/alzheimer_study'),
'out_preds': Path('runs/alzheimer_study/preds'),
'patience': 1,
'sample_idx_position': 0,
'save_pred_real_na': True}