Fit logistic regression model#
based on different imputation methods
baseline: reference
model: any other selected imputation method
Parameters#
Default and set parameters for the notebook.
folder_data: str = '' # specify data directory if needed
fn_clinical_data = "data/ALD_study/processed/ald_metadata_cli.csv"
folder_experiment = "runs/appl_ald_data/plasma/proteinGroups"
model_key = 'VAE'
target = 'kleiner'
sample_id_col = 'Sample ID'
cutoff_target: int = 2 # => for binarization target >= cutoff_target
file_format = "csv"
out_folder = 'diff_analysis'
fn_qc_samples = '' # 'data/ALD_study/processed/qc_plasma_proteinGroups.pkl'
baseline = 'RSN' # default is RSN, as this was used in the original ALD Niu. et. al 2022
template_pred = 'pred_real_na_{}.csv' # fixed, do not change
# Parameters
cutoff_target = 0.5
folder_experiment = "runs/alzheimer_study"
target = "AD"
baseline = "PI"
model_key = "DAE"
out_folder = "diff_analysis"
fn_clinical_data = "runs/alzheimer_study/data/clinical_data.csv"
root - INFO Removed from global namespace: folder_data
root - INFO Removed from global namespace: fn_clinical_data
root - INFO Removed from global namespace: folder_experiment
root - INFO Removed from global namespace: model_key
root - INFO Removed from global namespace: target
root - INFO Removed from global namespace: sample_id_col
root - INFO Removed from global namespace: cutoff_target
root - INFO Removed from global namespace: file_format
root - INFO Removed from global namespace: out_folder
root - INFO Removed from global namespace: fn_qc_samples
root - INFO Removed from global namespace: baseline
root - INFO Removed from global namespace: template_pred
root - INFO Already set attribute: folder_experiment has value runs/alzheimer_study
root - INFO Already set attribute: out_folder has value diff_analysis
{'baseline': 'PI',
'cutoff_target': 0.5,
'data': PosixPath('runs/alzheimer_study/data'),
'file_format': 'csv',
'fn_clinical_data': 'runs/alzheimer_study/data/clinical_data.csv',
'fn_qc_samples': '',
'folder_data': '',
'folder_experiment': PosixPath('runs/alzheimer_study'),
'model_key': 'DAE',
'out_figures': PosixPath('runs/alzheimer_study/figures'),
'out_folder': PosixPath('runs/alzheimer_study/diff_analysis/AD/PI_vs_DAE'),
'out_metrics': PosixPath('runs/alzheimer_study'),
'out_models': PosixPath('runs/alzheimer_study'),
'out_preds': PosixPath('runs/alzheimer_study/preds'),
'sample_id_col': 'Sample ID',
'target': 'AD',
'template_pred': 'pred_real_na_{}.csv'}
Load data#
Load target#
target = pd.read_csv(args.fn_clinical_data,
index_col=0,
usecols=[args.sample_id_col, args.target])
target = target.dropna()
target
| AD | |
|---|---|
| Sample ID | |
| Sample_000 | 0 |
| Sample_001 | 1 |
| Sample_002 | 1 |
| Sample_003 | 1 |
| Sample_004 | 1 |
| ... | ... |
| Sample_205 | 1 |
| Sample_206 | 0 |
| Sample_207 | 0 |
| Sample_208 | 0 |
| Sample_209 | 0 |
210 rows × 1 columns
MS proteomics or specified omics data#
Aggregated from data splits of the imputation workflow run before.
pimmslearn.io.datasplits - INFO Loaded 'train_X' from file: runs/alzheimer_study/data/train_X.csv
pimmslearn.io.datasplits - INFO Loaded 'val_y' from file: runs/alzheimer_study/data/val_y.csv
pimmslearn.io.datasplits - INFO Loaded 'test_y' from file: runs/alzheimer_study/data/test_y.csv
Sample ID protein groups
Sample_203 Q4KWH8;Q4KWH8-2;Q4KWH8-3;Q4KWH8-4 17.338
Sample_175 A0A087X1J7;P22352 19.835
Sample_037 K7ERI9;P02654 19.801
Sample_136 Q8N6C5;Q8N6C5-2;Q8N6C5-4 15.430
Sample_102 O75084;Q14332 14.926
Name: intensity, dtype: float64
Get overlap between independent features and target
Select by ALD criteria#
Use parameters as specified in ALD study.
root - INFO Initally: N samples: 210, M feat: 1421
root - INFO Dropped features quantified in less than 126 samples.
root - INFO After feat selection: N samples: 210, M feat: 1213
root - INFO Min No. of Protein-Groups in single sample: 754
root - INFO Finally: N samples: 210, M feat: 1213
| protein groups | A0A024QZX5;A0A087X1N8;P35237 | A0A024R0T9;K7ER74;P02655 | A0A024R3W6;A0A024R412;O60462;O60462-2;O60462-3;O60462-4;O60462-5;Q7LBX6;X5D2Q8 | A0A024R644;A0A0A0MRU5;A0A1B0GWI2;O75503 | A0A075B6H9 | A0A075B6I0 | A0A075B6I1 | A0A075B6I6 | A0A075B6I9 | A0A075B6J9 | ... | Q9Y653;Q9Y653-2;Q9Y653-3 | Q9Y696 | Q9Y6C2 | Q9Y6N6 | Q9Y6N7;Q9Y6N7-2;Q9Y6N7-4 | Q9Y6R7 | Q9Y6X5 | Q9Y6Y8;Q9Y6Y8-2 | Q9Y6Y9 | S4R3U6 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sample ID | |||||||||||||||||||||
| Sample_000 | 15.912 | 16.852 | 15.570 | 16.481 | 20.246 | 16.764 | 17.584 | 16.988 | 20.054 | NaN | ... | 16.012 | 15.178 | NaN | 15.050 | 16.842 | 19.863 | NaN | 19.563 | 12.837 | 12.805 |
| Sample_001 | 15.936 | 16.874 | 15.519 | 16.387 | 19.941 | 18.786 | 17.144 | NaN | 19.067 | 16.188 | ... | 15.528 | 15.576 | NaN | 14.833 | 16.597 | 20.299 | 15.556 | 19.386 | 13.970 | 12.442 |
| Sample_002 | 16.111 | 14.523 | 15.935 | 16.416 | 19.251 | 16.832 | 15.671 | 17.012 | 18.569 | NaN | ... | 15.229 | 14.728 | 13.757 | 15.118 | 17.440 | 19.598 | 15.735 | 20.447 | 12.636 | 12.505 |
| Sample_003 | 16.107 | 17.032 | 15.802 | 16.979 | 19.628 | 17.852 | 18.877 | 14.182 | 18.985 | 13.438 | ... | 15.495 | 14.590 | 14.682 | 15.140 | 17.356 | 19.429 | NaN | 20.216 | 12.627 | 12.445 |
| Sample_004 | 15.603 | 15.331 | 15.375 | 16.679 | 20.450 | 18.682 | 17.081 | 14.140 | 19.686 | 14.495 | ... | 14.757 | 15.094 | 14.048 | 15.256 | 17.075 | 19.582 | 15.328 | 19.867 | 13.145 | 12.235 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| Sample_205 | 15.682 | 16.886 | 14.910 | 16.482 | 17.705 | 17.039 | NaN | 16.413 | 19.102 | 16.064 | ... | 15.235 | 15.684 | 14.236 | 15.415 | 17.551 | 17.922 | 16.340 | 19.928 | 12.929 | 11.802 |
| Sample_206 | 15.798 | 17.554 | 15.600 | 15.938 | 18.154 | 18.152 | 16.503 | 16.860 | 18.538 | 15.288 | ... | 15.422 | 16.106 | NaN | 15.345 | 17.084 | 18.708 | 14.249 | 19.433 | NaN | NaN |
| Sample_207 | 15.739 | 16.877 | 15.469 | 16.898 | 18.636 | 17.950 | 16.321 | 16.401 | 18.849 | 17.580 | ... | 15.808 | 16.098 | 14.403 | 15.715 | 16.586 | 18.725 | 16.138 | 19.599 | 13.637 | 11.174 |
| Sample_208 | 15.477 | 16.779 | 14.995 | 16.132 | 14.908 | 17.530 | NaN | 16.119 | 18.368 | 15.202 | ... | 15.157 | 16.712 | NaN | 14.640 | 16.533 | 19.411 | 15.807 | 19.545 | 13.216 | NaN |
| Sample_209 | 15.727 | 17.261 | 15.175 | 16.235 | 17.893 | 17.744 | 16.371 | 15.780 | 18.806 | 16.532 | ... | 15.237 | 15.652 | 15.211 | 14.205 | 16.749 | 19.275 | 15.732 | 19.577 | 11.042 | 11.791 |
210 rows × 1213 columns
Number of complete cases which can be used:
Samples available both in proteomics data and for target: 210
Load imputations from specified model#
missing values pred. by DAE: runs/alzheimer_study/preds/pred_real_na_DAE.csv
Sample ID protein groups
Sample_004 P31151 14.881
Sample_135 A0A0A0MS09;P01880;P01880-2 17.023
Sample_068 P51674;P51674-2;P51674-3 14.009
Name: intensity, dtype: float64
Load imputations from baseline model#
Sample ID protein groups
Sample_000 A0A075B6J9 13.371
A0A075B6Q5 13.526
A0A075B6R2 12.825
A0A075B6S5 13.046
A0A087WSY4 13.247
...
Sample_209 Q9P1W8;Q9P1W8-2;Q9P1W8-4 13.651
Q9UI40;Q9UI40-2 12.912
Q9UIW2 13.541
Q9UMX0;Q9UMX0-2;Q9UMX0-4 12.938
Q9UP79 13.357
Name: intensity, Length: 46401, dtype: float64
Modeling setup#
General approach:
use one train, test split of the data
select best 10 features from training data
X_train,y_trainbefore binarization of targetdichotomize (binarize) data into to groups (zero and 1)
evaluate model on the test data
X_test,y_test
Repeat general approach for
all original ald data: all features justed in original ALD study
all model data: all features available my using the self supervised deep learning model
newly available feat only: the subset of features available from the self supervised deep learning model which were newly retained using the new approach
All data:
| protein groups | A0A024QZX5;A0A087X1N8;P35237 | A0A024R0T9;K7ER74;P02655 | A0A024R3W6;A0A024R412;O60462;O60462-2;O60462-3;O60462-4;O60462-5;Q7LBX6;X5D2Q8 | A0A024R644;A0A0A0MRU5;A0A1B0GWI2;O75503 | A0A075B6H7 | A0A075B6H9 | A0A075B6I0 | A0A075B6I1 | A0A075B6I6 | A0A075B6I9 | ... | Q9Y653;Q9Y653-2;Q9Y653-3 | Q9Y696 | Q9Y6C2 | Q9Y6N6 | Q9Y6N7;Q9Y6N7-2;Q9Y6N7-4 | Q9Y6R7 | Q9Y6X5 | Q9Y6Y8;Q9Y6Y8-2 | Q9Y6Y9 | S4R3U6 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sample ID | |||||||||||||||||||||
| Sample_000 | 15.912 | 16.852 | 15.570 | 16.481 | 17.301 | 20.246 | 16.764 | 17.584 | 16.988 | 20.054 | ... | 16.012 | 15.178 | 14.268 | 15.050 | 16.842 | 19.863 | 15.936 | 19.563 | 12.837 | 12.805 |
| Sample_001 | 15.936 | 16.874 | 15.519 | 16.387 | 13.796 | 19.941 | 18.786 | 17.144 | 16.917 | 19.067 | ... | 15.528 | 15.576 | 14.329 | 14.833 | 16.597 | 20.299 | 15.556 | 19.386 | 13.970 | 12.442 |
| Sample_002 | 16.111 | 14.523 | 15.935 | 16.416 | 18.175 | 19.251 | 16.832 | 15.671 | 17.012 | 18.569 | ... | 15.229 | 14.728 | 13.757 | 15.118 | 17.440 | 19.598 | 15.735 | 20.447 | 12.636 | 12.505 |
| Sample_003 | 16.107 | 17.032 | 15.802 | 16.979 | 15.963 | 19.628 | 17.852 | 18.877 | 14.182 | 18.985 | ... | 15.495 | 14.590 | 14.682 | 15.140 | 17.356 | 19.429 | 15.719 | 20.216 | 12.627 | 12.445 |
| Sample_004 | 15.603 | 15.331 | 15.375 | 16.679 | 15.473 | 20.450 | 18.682 | 17.081 | 14.140 | 19.686 | ... | 14.757 | 15.094 | 14.048 | 15.256 | 17.075 | 19.582 | 15.328 | 19.867 | 13.145 | 12.235 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| Sample_205 | 15.682 | 16.886 | 14.910 | 16.482 | 14.969 | 17.705 | 17.039 | 15.528 | 16.413 | 19.102 | ... | 15.235 | 15.684 | 14.236 | 15.415 | 17.551 | 17.922 | 16.340 | 19.928 | 12.929 | 11.802 |
| Sample_206 | 15.798 | 17.554 | 15.600 | 15.938 | 15.219 | 18.154 | 18.152 | 16.503 | 16.860 | 18.538 | ... | 15.422 | 16.106 | 14.605 | 15.345 | 17.084 | 18.708 | 14.249 | 19.433 | 11.927 | 10.992 |
| Sample_207 | 15.739 | 16.877 | 15.469 | 16.898 | 13.733 | 18.636 | 17.950 | 16.321 | 16.401 | 18.849 | ... | 15.808 | 16.098 | 14.403 | 15.715 | 16.586 | 18.725 | 16.138 | 19.599 | 13.637 | 11.174 |
| Sample_208 | 15.477 | 16.779 | 14.995 | 16.132 | 13.486 | 14.908 | 17.530 | 16.394 | 16.119 | 18.368 | ... | 15.157 | 16.712 | 14.376 | 14.640 | 16.533 | 19.411 | 15.807 | 19.545 | 13.216 | 11.095 |
| Sample_209 | 15.727 | 17.261 | 15.175 | 16.235 | 14.686 | 17.893 | 17.744 | 16.371 | 15.780 | 18.806 | ... | 15.237 | 15.652 | 15.211 | 14.205 | 16.749 | 19.275 | 15.732 | 19.577 | 11.042 | 11.791 |
210 rows × 1421 columns
Subset of data by ALD criteria#
| protein groups | A0A024QZX5;A0A087X1N8;P35237 | A0A024R0T9;K7ER74;P02655 | A0A024R3W6;A0A024R412;O60462;O60462-2;O60462-3;O60462-4;O60462-5;Q7LBX6;X5D2Q8 | A0A024R644;A0A0A0MRU5;A0A1B0GWI2;O75503 | A0A075B6H9 | A0A075B6I0 | A0A075B6I1 | A0A075B6I6 | A0A075B6I9 | A0A075B6K4 | ... | O14793 | O95479;R4GMU1 | P01282;P01282-2 | P10619;P10619-2;X6R5C5;X6R8A1 | P21810 | Q14956;Q14956-2 | Q6ZMP0;Q6ZMP0-2 | Q9HBW1 | Q9NY15 | P17050 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sample ID | |||||||||||||||||||||
| Sample_000 | 15.912 | 16.852 | 15.570 | 16.481 | 20.246 | 16.764 | 17.584 | 16.988 | 20.054 | 16.148 | ... | 13.823 | 12.710 | 12.289 | 12.429 | 12.961 | 14.060 | 13.097 | 13.328 | 13.146 | 12.471 |
| Sample_001 | 15.936 | 16.874 | 15.519 | 16.387 | 19.941 | 18.786 | 17.144 | 12.498 | 19.067 | 16.127 | ... | 12.419 | 11.977 | 13.384 | 14.106 | 13.555 | 14.165 | 11.243 | 14.072 | 13.591 | 12.031 |
| Sample_002 | 16.111 | 14.523 | 15.935 | 16.416 | 19.251 | 16.832 | 15.671 | 17.012 | 18.569 | 15.387 | ... | 12.819 | 13.346 | 12.788 | 11.961 | 12.539 | 12.713 | 12.752 | 13.852 | 13.730 | 13.515 |
| Sample_003 | 16.107 | 17.032 | 15.802 | 16.979 | 19.628 | 17.852 | 18.877 | 14.182 | 18.985 | 16.565 | ... | 11.799 | 11.335 | 12.792 | 13.810 | 14.088 | 10.896 | 13.095 | 12.938 | 13.069 | 11.550 |
| Sample_004 | 15.603 | 15.331 | 15.375 | 16.679 | 20.450 | 18.682 | 17.081 | 14.140 | 19.686 | 16.418 | ... | 13.601 | 14.277 | 13.066 | 13.071 | 12.212 | 13.470 | 13.209 | 11.914 | 12.317 | 12.259 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| Sample_205 | 15.682 | 16.886 | 14.910 | 16.482 | 17.705 | 17.039 | 11.566 | 16.413 | 19.102 | 15.350 | ... | 14.269 | 14.064 | 16.826 | 18.182 | 15.225 | 15.044 | 14.192 | 16.605 | 14.995 | 14.257 |
| Sample_206 | 15.798 | 17.554 | 15.600 | 15.938 | 18.154 | 18.152 | 16.503 | 16.860 | 18.538 | 16.582 | ... | 14.273 | 17.700 | 16.802 | 20.202 | 15.280 | 15.086 | 13.978 | 18.086 | 15.557 | 14.171 |
| Sample_207 | 15.739 | 16.877 | 15.469 | 16.898 | 18.636 | 17.950 | 16.321 | 16.401 | 18.849 | 15.768 | ... | 14.473 | 16.882 | 16.917 | 20.105 | 15.690 | 15.135 | 13.138 | 17.066 | 15.706 | 15.690 |
| Sample_208 | 15.477 | 16.779 | 14.995 | 16.132 | 14.908 | 17.530 | 12.104 | 16.119 | 18.368 | 17.560 | ... | 15.234 | 17.175 | 16.521 | 18.859 | 15.305 | 15.161 | 13.006 | 17.917 | 15.396 | 14.371 |
| Sample_209 | 15.727 | 17.261 | 15.175 | 16.235 | 17.893 | 17.744 | 16.371 | 15.780 | 18.806 | 16.338 | ... | 14.556 | 16.656 | 16.954 | 18.493 | 15.823 | 14.626 | 13.385 | 17.767 | 15.687 | 13.573 |
210 rows × 1213 columns
Features which would not have been included using ALD criteria:
Index(['A0A075B6H7', 'A0A075B6Q5', 'A0A075B7B8', 'A0A087WSY4',
'A0A087WTT8;A0A0A0MQX5;O94779;O94779-2', 'A0A087WXB8;Q9Y274',
'A0A087WXE9;E9PQ70;Q6UXH9;Q6UXH9-2;Q6UXH9-3',
'A0A087X1Z2;C9JTV4;H0Y4Y4;Q8WYH2;Q96C19;Q9BUP0;Q9BUP0-2',
'A0A0A0MQS9;A0A0A0MTC7;Q16363;Q16363-2', 'A0A0A0MSN4;P12821;P12821-2',
...
'Q9NZ94;Q9NZ94-2;Q9NZ94-3', 'Q9NZU1', 'Q9P1W8;Q9P1W8-2;Q9P1W8-4',
'Q9UHI8', 'Q9UI40;Q9UI40-2',
'Q9UIB8;Q9UIB8-2;Q9UIB8-3;Q9UIB8-4;Q9UIB8-5;Q9UIB8-6',
'Q9UKZ4;Q9UKZ4-2', 'Q9UMX0;Q9UMX0-2;Q9UMX0-4', 'Q9Y281;Q9Y281-3',
'Q9Y490'],
dtype='object', name='protein groups', length=208)
Binarize targets, but also keep groups for stratification
| AD | 0 | 1 |
|---|---|---|
| AD | ||
| False | 122 | 0 |
| True | 0 | 88 |
Determine best number of parameters by cross validation procedure#
using subset of data by ALD criteria:
0%| | 0/1 [00:00<?, ?it/s]
100%|██████████| 1/1 [00:00<00:00, 124.44it/s]
0%| | 0/2 [00:00<?, ?it/s]
100%|██████████| 2/2 [00:00<00:00, 4.92it/s]
100%|██████████| 2/2 [00:00<00:00, 4.90it/s]
0%| | 0/3 [00:00<?, ?it/s]
67%|██████▋ | 2/3 [00:00<00:00, 5.39it/s]
100%|██████████| 3/3 [00:00<00:00, 3.56it/s]
100%|██████████| 3/3 [00:00<00:00, 3.80it/s]
0%| | 0/4 [00:00<?, ?it/s]
50%|█████ | 2/4 [00:00<00:00, 9.56it/s]
75%|███████▌ | 3/4 [00:00<00:00, 7.06it/s]
100%|██████████| 4/4 [00:00<00:00, 5.79it/s]
100%|██████████| 4/4 [00:00<00:00, 6.36it/s]
0%| | 0/5 [00:00<?, ?it/s]
40%|████ | 2/5 [00:00<00:00, 7.79it/s]
60%|██████ | 3/5 [00:00<00:00, 5.33it/s]
80%|████████ | 4/5 [00:00<00:00, 4.65it/s]
100%|██████████| 5/5 [00:01<00:00, 3.77it/s]
100%|██████████| 5/5 [00:01<00:00, 4.33it/s]
0%| | 0/6 [00:00<?, ?it/s]
33%|███▎ | 2/6 [00:00<00:00, 5.62it/s]
50%|█████ | 3/6 [00:00<00:00, 3.88it/s]
67%|██████▋ | 4/6 [00:01<00:00, 3.63it/s]
83%|████████▎ | 5/6 [00:01<00:00, 3.33it/s]
100%|██████████| 6/6 [00:01<00:00, 3.27it/s]
100%|██████████| 6/6 [00:01<00:00, 3.53it/s]
0%| | 0/7 [00:00<?, ?it/s]
29%|██▊ | 2/7 [00:00<00:00, 6.51it/s]
43%|████▎ | 3/7 [00:00<00:00, 5.01it/s]
57%|█████▋ | 4/7 [00:00<00:00, 4.32it/s]
71%|███████▏ | 5/7 [00:01<00:00, 4.01it/s]
86%|████████▌ | 6/7 [00:01<00:00, 3.19it/s]
100%|██████████| 7/7 [00:01<00:00, 3.06it/s]
100%|██████████| 7/7 [00:01<00:00, 3.61it/s]
0%| | 0/8 [00:00<?, ?it/s]
25%|██▌ | 2/8 [00:00<00:00, 9.67it/s]
38%|███▊ | 3/8 [00:00<00:00, 6.34it/s]
50%|█████ | 4/8 [00:00<00:00, 4.57it/s]
62%|██████▎ | 5/8 [00:01<00:00, 3.92it/s]
75%|███████▌ | 6/8 [00:01<00:00, 3.53it/s]
88%|████████▊ | 7/8 [00:01<00:00, 3.32it/s]
100%|██████████| 8/8 [00:02<00:00, 3.24it/s]
100%|██████████| 8/8 [00:02<00:00, 3.83it/s]
0%| | 0/9 [00:00<?, ?it/s]
22%|██▏ | 2/9 [00:00<00:01, 5.40it/s]
33%|███▎ | 3/9 [00:00<00:01, 4.42it/s]
44%|████▍ | 4/9 [00:00<00:01, 4.24it/s]
56%|█████▌ | 5/9 [00:01<00:01, 3.84it/s]
67%|██████▋ | 6/9 [00:01<00:00, 3.93it/s]
78%|███████▊ | 7/9 [00:01<00:00, 4.15it/s]
89%|████████▉ | 8/9 [00:01<00:00, 3.94it/s]
100%|██████████| 9/9 [00:02<00:00, 3.60it/s]
100%|██████████| 9/9 [00:02<00:00, 3.94it/s]
0%| | 0/10 [00:00<?, ?it/s]
20%|██ | 2/10 [00:00<00:01, 6.38it/s]
30%|███ | 3/10 [00:00<00:01, 4.96it/s]
40%|████ | 4/10 [00:00<00:01, 4.49it/s]
50%|█████ | 5/10 [00:01<00:01, 4.18it/s]
60%|██████ | 6/10 [00:01<00:01, 3.98it/s]
70%|███████ | 7/10 [00:01<00:00, 3.70it/s]
80%|████████ | 8/10 [00:01<00:00, 3.59it/s]
90%|█████████ | 9/10 [00:02<00:00, 3.41it/s]
100%|██████████| 10/10 [00:02<00:00, 3.43it/s]
100%|██████████| 10/10 [00:02<00:00, 3.84it/s]
0%| | 0/11 [00:00<?, ?it/s]
18%|█▊ | 2/11 [00:00<00:01, 7.23it/s]
27%|██▋ | 3/11 [00:00<00:01, 5.57it/s]
36%|███▋ | 4/11 [00:00<00:01, 4.71it/s]
45%|████▌ | 5/11 [00:01<00:01, 4.22it/s]
55%|█████▍ | 6/11 [00:01<00:01, 3.30it/s]
64%|██████▎ | 7/11 [00:01<00:01, 2.91it/s]
73%|███████▎ | 8/11 [00:02<00:01, 2.80it/s]
82%|████████▏ | 9/11 [00:02<00:00, 2.56it/s]
91%|█████████ | 10/11 [00:03<00:00, 2.72it/s]
100%|██████████| 11/11 [00:03<00:00, 2.78it/s]
100%|██████████| 11/11 [00:03<00:00, 3.20it/s]
0%| | 0/12 [00:00<?, ?it/s]
17%|█▋ | 2/12 [00:00<00:01, 5.85it/s]
25%|██▌ | 3/12 [00:00<00:02, 4.24it/s]
33%|███▎ | 4/12 [00:00<00:01, 4.15it/s]
42%|████▏ | 5/12 [00:01<00:02, 3.45it/s]
50%|█████ | 6/12 [00:01<00:01, 3.13it/s]
58%|█████▊ | 7/12 [00:02<00:01, 2.84it/s]
67%|██████▋ | 8/12 [00:02<00:01, 2.85it/s]
75%|███████▌ | 9/12 [00:02<00:01, 2.97it/s]
83%|████████▎ | 10/12 [00:03<00:00, 2.79it/s]
92%|█████████▏| 11/12 [00:03<00:00, 2.97it/s]
100%|██████████| 12/12 [00:03<00:00, 3.18it/s]
100%|██████████| 12/12 [00:03<00:00, 3.23it/s]
0%| | 0/13 [00:00<?, ?it/s]
15%|█▌ | 2/13 [00:00<00:01, 8.55it/s]
23%|██▎ | 3/13 [00:00<00:01, 6.02it/s]
31%|███ | 4/13 [00:00<00:01, 5.21it/s]
38%|███▊ | 5/13 [00:00<00:01, 4.83it/s]
46%|████▌ | 6/13 [00:01<00:01, 4.56it/s]
54%|█████▍ | 7/13 [00:01<00:01, 4.48it/s]
62%|██████▏ | 8/13 [00:01<00:01, 4.47it/s]
69%|██████▉ | 9/13 [00:01<00:00, 4.32it/s]
77%|███████▋ | 10/13 [00:02<00:00, 3.93it/s]
85%|████████▍ | 11/13 [00:02<00:00, 3.97it/s]
92%|█████████▏| 12/13 [00:02<00:00, 3.28it/s]
100%|██████████| 13/13 [00:03<00:00, 3.11it/s]
100%|██████████| 13/13 [00:03<00:00, 4.02it/s]
0%| | 0/14 [00:00<?, ?it/s]
14%|█▍ | 2/14 [00:00<00:01, 8.46it/s]
21%|██▏ | 3/14 [00:00<00:01, 5.85it/s]
29%|██▊ | 4/14 [00:00<00:01, 5.36it/s]
36%|███▌ | 5/14 [00:00<00:01, 4.79it/s]
43%|████▎ | 6/14 [00:01<00:02, 3.39it/s]
50%|█████ | 7/14 [00:01<00:02, 3.01it/s]
57%|█████▋ | 8/14 [00:02<00:02, 2.88it/s]
64%|██████▍ | 9/14 [00:02<00:01, 2.71it/s]
71%|███████▏ | 10/14 [00:02<00:01, 2.92it/s]
79%|███████▊ | 11/14 [00:03<00:00, 3.11it/s]
86%|████████▌ | 12/14 [00:03<00:00, 3.36it/s]
93%|█████████▎| 13/14 [00:03<00:00, 3.50it/s]
100%|██████████| 14/14 [00:03<00:00, 3.53it/s]
100%|██████████| 14/14 [00:03<00:00, 3.54it/s]
0%| | 0/15 [00:00<?, ?it/s]
13%|█▎ | 2/15 [00:00<00:02, 5.16it/s]
20%|██ | 3/15 [00:00<00:03, 3.71it/s]
27%|██▋ | 4/15 [00:01<00:03, 3.56it/s]
33%|███▎ | 5/15 [00:01<00:02, 3.58it/s]
40%|████ | 6/15 [00:01<00:02, 3.56it/s]
47%|████▋ | 7/15 [00:01<00:02, 3.51it/s]
53%|█████▎ | 8/15 [00:02<00:02, 3.42it/s]
60%|██████ | 9/15 [00:02<00:01, 3.44it/s]
67%|██████▋ | 10/15 [00:02<00:01, 3.51it/s]
73%|███████▎ | 11/15 [00:03<00:01, 3.71it/s]
80%|████████ | 12/15 [00:03<00:00, 3.74it/s]
87%|████████▋ | 13/15 [00:03<00:00, 3.92it/s]
93%|█████████▎| 14/15 [00:03<00:00, 4.07it/s]
100%|██████████| 15/15 [00:04<00:00, 3.60it/s]
100%|██████████| 15/15 [00:04<00:00, 3.67it/s]
| fit_time | score_time | test_precision | test_recall | test_f1 | test_balanced_accuracy | test_roc_auc | test_average_precision | n_observations | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| mean | std | mean | std | mean | std | mean | std | mean | std | mean | std | mean | std | mean | std | mean | std | |
| n_features | ||||||||||||||||||
| 1 | 0.004 | 0.002 | 0.049 | 0.019 | 0.691 | 0.406 | 0.099 | 0.086 | 0.165 | 0.133 | 0.542 | 0.042 | 0.855 | 0.062 | 0.827 | 0.087 | 210.000 | 0.000 |
| 2 | 0.004 | 0.002 | 0.045 | 0.016 | 0.707 | 0.089 | 0.600 | 0.103 | 0.643 | 0.079 | 0.707 | 0.058 | 0.788 | 0.066 | 0.762 | 0.084 | 210.000 | 0.000 |
| 3 | 0.005 | 0.003 | 0.060 | 0.023 | 0.720 | 0.084 | 0.654 | 0.114 | 0.679 | 0.082 | 0.732 | 0.060 | 0.804 | 0.066 | 0.770 | 0.087 | 210.000 | 0.000 |
| 4 | 0.006 | 0.003 | 0.060 | 0.023 | 0.721 | 0.086 | 0.679 | 0.110 | 0.693 | 0.074 | 0.741 | 0.057 | 0.799 | 0.063 | 0.762 | 0.086 | 210.000 | 0.000 |
| 5 | 0.004 | 0.001 | 0.043 | 0.015 | 0.713 | 0.089 | 0.665 | 0.112 | 0.681 | 0.078 | 0.732 | 0.058 | 0.794 | 0.063 | 0.762 | 0.084 | 210.000 | 0.000 |
| 6 | 0.004 | 0.002 | 0.046 | 0.016 | 0.732 | 0.084 | 0.700 | 0.105 | 0.710 | 0.070 | 0.754 | 0.054 | 0.811 | 0.064 | 0.787 | 0.083 | 210.000 | 0.000 |
| 7 | 0.006 | 0.003 | 0.054 | 0.023 | 0.788 | 0.087 | 0.819 | 0.100 | 0.800 | 0.076 | 0.828 | 0.065 | 0.894 | 0.049 | 0.869 | 0.059 | 210.000 | 0.000 |
| 8 | 0.006 | 0.004 | 0.062 | 0.027 | 0.789 | 0.088 | 0.822 | 0.101 | 0.801 | 0.076 | 0.829 | 0.065 | 0.892 | 0.050 | 0.866 | 0.063 | 210.000 | 0.000 |
| 9 | 0.005 | 0.002 | 0.055 | 0.020 | 0.788 | 0.081 | 0.816 | 0.097 | 0.798 | 0.072 | 0.827 | 0.062 | 0.896 | 0.051 | 0.873 | 0.060 | 210.000 | 0.000 |
| 10 | 0.005 | 0.002 | 0.055 | 0.020 | 0.809 | 0.074 | 0.818 | 0.091 | 0.811 | 0.066 | 0.838 | 0.056 | 0.912 | 0.047 | 0.901 | 0.049 | 210.000 | 0.000 |
| 11 | 0.004 | 0.002 | 0.041 | 0.011 | 0.813 | 0.078 | 0.819 | 0.084 | 0.814 | 0.066 | 0.840 | 0.057 | 0.913 | 0.047 | 0.903 | 0.049 | 210.000 | 0.000 |
| 12 | 0.006 | 0.003 | 0.057 | 0.024 | 0.819 | 0.080 | 0.822 | 0.088 | 0.818 | 0.069 | 0.844 | 0.059 | 0.912 | 0.047 | 0.900 | 0.049 | 210.000 | 0.000 |
| 13 | 0.005 | 0.003 | 0.050 | 0.018 | 0.849 | 0.084 | 0.806 | 0.087 | 0.824 | 0.067 | 0.849 | 0.056 | 0.918 | 0.044 | 0.909 | 0.045 | 210.000 | 0.000 |
| 14 | 0.004 | 0.001 | 0.040 | 0.009 | 0.843 | 0.088 | 0.805 | 0.089 | 0.820 | 0.067 | 0.846 | 0.057 | 0.915 | 0.045 | 0.907 | 0.046 | 210.000 | 0.000 |
| 15 | 0.005 | 0.002 | 0.048 | 0.019 | 0.830 | 0.089 | 0.807 | 0.089 | 0.814 | 0.068 | 0.841 | 0.058 | 0.915 | 0.045 | 0.907 | 0.045 | 210.000 | 0.000 |
Using all data:
0%| | 0/1 [00:00<?, ?it/s]
100%|██████████| 1/1 [00:00<00:00, 288.03it/s]
0%| | 0/2 [00:00<?, ?it/s]
100%|██████████| 2/2 [00:00<00:00, 5.82it/s]
100%|██████████| 2/2 [00:00<00:00, 5.79it/s]
0%| | 0/3 [00:00<?, ?it/s]
67%|██████▋ | 2/3 [00:00<00:00, 7.53it/s]
100%|██████████| 3/3 [00:00<00:00, 5.22it/s]
100%|██████████| 3/3 [00:00<00:00, 5.54it/s]
0%| | 0/4 [00:00<?, ?it/s]
50%|█████ | 2/4 [00:00<00:00, 4.98it/s]
75%|███████▌ | 3/4 [00:00<00:00, 3.93it/s]
100%|██████████| 4/4 [00:01<00:00, 3.57it/s]
100%|██████████| 4/4 [00:01<00:00, 3.78it/s]
0%| | 0/5 [00:00<?, ?it/s]
40%|████ | 2/5 [00:00<00:00, 3.83it/s]
60%|██████ | 3/5 [00:00<00:00, 2.90it/s]
80%|████████ | 4/5 [00:01<00:00, 2.62it/s]
100%|██████████| 5/5 [00:01<00:00, 2.53it/s]
100%|██████████| 5/5 [00:01<00:00, 2.70it/s]
0%| | 0/6 [00:00<?, ?it/s]
33%|███▎ | 2/6 [00:00<00:00, 8.04it/s]
50%|█████ | 3/6 [00:00<00:00, 4.14it/s]
67%|██████▋ | 4/6 [00:01<00:00, 3.14it/s]
83%|████████▎ | 5/6 [00:01<00:00, 2.84it/s]
100%|██████████| 6/6 [00:02<00:00, 2.46it/s]
100%|██████████| 6/6 [00:02<00:00, 2.95it/s]
0%| | 0/7 [00:00<?, ?it/s]
29%|██▊ | 2/7 [00:00<00:00, 6.13it/s]
43%|████▎ | 3/7 [00:00<00:00, 4.46it/s]
57%|█████▋ | 4/7 [00:00<00:00, 3.98it/s]
71%|███████▏ | 5/7 [00:01<00:00, 4.01it/s]
86%|████████▌ | 6/7 [00:01<00:00, 4.01it/s]
100%|██████████| 7/7 [00:01<00:00, 3.98it/s]
100%|██████████| 7/7 [00:01<00:00, 4.16it/s]
0%| | 0/8 [00:00<?, ?it/s]
25%|██▌ | 2/8 [00:00<00:01, 5.55it/s]
38%|███▊ | 3/8 [00:00<00:01, 3.29it/s]
50%|█████ | 4/8 [00:01<00:01, 2.33it/s]
62%|██████▎ | 5/8 [00:01<00:01, 2.31it/s]
75%|███████▌ | 6/8 [00:02<00:00, 2.28it/s]
88%|████████▊ | 7/8 [00:02<00:00, 2.24it/s]
100%|██████████| 8/8 [00:03<00:00, 2.34it/s]
100%|██████████| 8/8 [00:03<00:00, 2.49it/s]
0%| | 0/9 [00:00<?, ?it/s]
22%|██▏ | 2/9 [00:00<00:01, 4.10it/s]
33%|███▎ | 3/9 [00:00<00:02, 2.94it/s]
44%|████▍ | 4/9 [00:01<00:01, 3.04it/s]
56%|█████▌ | 5/9 [00:01<00:01, 2.90it/s]
67%|██████▋ | 6/9 [00:02<00:01, 2.80it/s]
78%|███████▊ | 7/9 [00:02<00:00, 2.67it/s]
89%|████████▉ | 8/9 [00:02<00:00, 2.87it/s]
100%|██████████| 9/9 [00:03<00:00, 2.96it/s]
100%|██████████| 9/9 [00:03<00:00, 2.95it/s]
0%| | 0/10 [00:00<?, ?it/s]
20%|██ | 2/10 [00:00<00:01, 4.49it/s]
30%|███ | 3/10 [00:00<00:01, 3.54it/s]
40%|████ | 4/10 [00:01<00:01, 3.24it/s]
50%|█████ | 5/10 [00:01<00:01, 3.10it/s]
60%|██████ | 6/10 [00:01<00:01, 3.34it/s]
70%|███████ | 7/10 [00:02<00:00, 3.37it/s]
80%|████████ | 8/10 [00:02<00:00, 3.37it/s]
90%|█████████ | 9/10 [00:02<00:00, 3.40it/s]
100%|██████████| 10/10 [00:02<00:00, 3.39it/s]
100%|██████████| 10/10 [00:02<00:00, 3.40it/s]
0%| | 0/11 [00:00<?, ?it/s]
18%|█▊ | 2/11 [00:00<00:01, 7.51it/s]
27%|██▋ | 3/11 [00:00<00:01, 5.48it/s]
36%|███▋ | 4/11 [00:00<00:01, 4.59it/s]
45%|████▌ | 5/11 [00:01<00:01, 4.35it/s]
55%|█████▍ | 6/11 [00:01<00:01, 3.49it/s]
64%|██████▎ | 7/11 [00:01<00:01, 3.58it/s]
73%|███████▎ | 8/11 [00:01<00:00, 3.74it/s]
82%|████████▏ | 9/11 [00:02<00:00, 3.79it/s]
91%|█████████ | 10/11 [00:02<00:00, 3.88it/s]
100%|██████████| 11/11 [00:02<00:00, 3.98it/s]
100%|██████████| 11/11 [00:02<00:00, 4.09it/s]
0%| | 0/12 [00:00<?, ?it/s]
17%|█▋ | 2/12 [00:00<00:01, 7.85it/s]
25%|██▌ | 3/12 [00:00<00:01, 5.54it/s]
33%|███▎ | 4/12 [00:00<00:01, 4.92it/s]
42%|████▏ | 5/12 [00:00<00:01, 4.60it/s]
50%|█████ | 6/12 [00:01<00:01, 3.76it/s]
58%|█████▊ | 7/12 [00:01<00:01, 3.64it/s]
67%|██████▋ | 8/12 [00:01<00:01, 3.53it/s]
75%|███████▌ | 9/12 [00:02<00:01, 2.93it/s]
83%|████████▎ | 10/12 [00:02<00:00, 2.77it/s]
92%|█████████▏| 11/12 [00:03<00:00, 2.74it/s]
100%|██████████| 12/12 [00:03<00:00, 2.86it/s]
100%|██████████| 12/12 [00:03<00:00, 3.41it/s]
0%| | 0/13 [00:00<?, ?it/s]
15%|█▌ | 2/13 [00:00<00:01, 6.78it/s]
23%|██▎ | 3/13 [00:00<00:01, 5.01it/s]
31%|███ | 4/13 [00:00<00:02, 4.12it/s]
38%|███▊ | 5/13 [00:01<00:02, 3.96it/s]
46%|████▌ | 6/13 [00:01<00:02, 3.31it/s]
54%|█████▍ | 7/13 [00:01<00:01, 3.07it/s]
62%|██████▏ | 8/13 [00:02<00:01, 2.81it/s]
69%|██████▉ | 9/13 [00:02<00:01, 2.67it/s]
77%|███████▋ | 10/13 [00:03<00:01, 2.85it/s]
85%|████████▍ | 11/13 [00:03<00:00, 3.06it/s]
92%|█████████▏| 12/13 [00:03<00:00, 3.03it/s]
100%|██████████| 13/13 [00:03<00:00, 3.11it/s]
100%|██████████| 13/13 [00:03<00:00, 3.26it/s]
0%| | 0/14 [00:00<?, ?it/s]
14%|█▍ | 2/14 [00:00<00:02, 4.15it/s]
21%|██▏ | 3/14 [00:00<00:03, 3.11it/s]
29%|██▊ | 4/14 [00:01<00:03, 2.89it/s]
36%|███▌ | 5/14 [00:01<00:03, 2.95it/s]
43%|████▎ | 6/14 [00:01<00:02, 3.09it/s]
50%|█████ | 7/14 [00:02<00:02, 3.26it/s]
57%|█████▋ | 8/14 [00:02<00:01, 3.41it/s]
64%|██████▍ | 9/14 [00:02<00:01, 3.60it/s]
71%|███████▏ | 10/14 [00:02<00:01, 3.61it/s]
79%|███████▊ | 11/14 [00:03<00:00, 3.72it/s]
86%|████████▌ | 12/14 [00:03<00:00, 3.65it/s]
93%|█████████▎| 13/14 [00:03<00:00, 3.37it/s]
100%|██████████| 14/14 [00:04<00:00, 3.45it/s]
100%|██████████| 14/14 [00:04<00:00, 3.38it/s]
0%| | 0/15 [00:00<?, ?it/s]
13%|█▎ | 2/15 [00:00<00:02, 6.15it/s]
20%|██ | 3/15 [00:00<00:02, 4.68it/s]
27%|██▋ | 4/15 [00:00<00:02, 4.27it/s]
33%|███▎ | 5/15 [00:01<00:02, 3.99it/s]
40%|████ | 6/15 [00:01<00:02, 3.79it/s]
47%|████▋ | 7/15 [00:01<00:02, 3.65it/s]
53%|█████▎ | 8/15 [00:02<00:01, 3.66it/s]
60%|██████ | 9/15 [00:02<00:01, 3.58it/s]
67%|██████▋ | 10/15 [00:02<00:01, 3.61it/s]
73%|███████▎ | 11/15 [00:02<00:01, 3.19it/s]
80%|████████ | 12/15 [00:03<00:00, 3.29it/s]
87%|████████▋ | 13/15 [00:03<00:00, 3.45it/s]
93%|█████████▎| 14/15 [00:03<00:00, 3.51it/s]
100%|██████████| 15/15 [00:04<00:00, 3.64it/s]
100%|██████████| 15/15 [00:04<00:00, 3.71it/s]
| fit_time | score_time | test_precision | test_recall | test_f1 | test_balanced_accuracy | test_roc_auc | test_average_precision | n_observations | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| mean | std | mean | std | mean | std | mean | std | mean | std | mean | std | mean | std | mean | std | mean | std | |
| n_features | ||||||||||||||||||
| 1 | 0.004 | 0.001 | 0.041 | 0.013 | 0.194 | 0.350 | 0.023 | 0.049 | 0.040 | 0.080 | 0.506 | 0.020 | 0.867 | 0.059 | 0.833 | 0.086 | 210.000 | 0.000 |
| 2 | 0.004 | 0.002 | 0.043 | 0.017 | 0.643 | 0.101 | 0.550 | 0.129 | 0.585 | 0.097 | 0.662 | 0.066 | 0.719 | 0.076 | 0.677 | 0.085 | 210.000 | 0.000 |
| 3 | 0.005 | 0.003 | 0.050 | 0.018 | 0.750 | 0.089 | 0.608 | 0.126 | 0.663 | 0.090 | 0.728 | 0.064 | 0.778 | 0.068 | 0.761 | 0.077 | 210.000 | 0.000 |
| 4 | 0.004 | 0.002 | 0.047 | 0.019 | 0.750 | 0.094 | 0.608 | 0.121 | 0.664 | 0.090 | 0.729 | 0.065 | 0.772 | 0.068 | 0.757 | 0.077 | 210.000 | 0.000 |
| 5 | 0.004 | 0.001 | 0.045 | 0.015 | 0.722 | 0.097 | 0.582 | 0.117 | 0.638 | 0.092 | 0.708 | 0.066 | 0.778 | 0.066 | 0.752 | 0.079 | 210.000 | 0.000 |
| 6 | 0.005 | 0.002 | 0.054 | 0.018 | 0.702 | 0.083 | 0.611 | 0.115 | 0.648 | 0.084 | 0.711 | 0.060 | 0.782 | 0.064 | 0.748 | 0.078 | 210.000 | 0.000 |
| 7 | 0.006 | 0.003 | 0.068 | 0.026 | 0.700 | 0.092 | 0.609 | 0.118 | 0.644 | 0.085 | 0.708 | 0.061 | 0.779 | 0.064 | 0.747 | 0.079 | 210.000 | 0.000 |
| 8 | 0.005 | 0.002 | 0.058 | 0.022 | 0.745 | 0.088 | 0.623 | 0.115 | 0.673 | 0.084 | 0.733 | 0.060 | 0.819 | 0.062 | 0.779 | 0.088 | 210.000 | 0.000 |
| 9 | 0.004 | 0.002 | 0.045 | 0.014 | 0.785 | 0.087 | 0.807 | 0.100 | 0.791 | 0.071 | 0.821 | 0.061 | 0.891 | 0.052 | 0.854 | 0.070 | 210.000 | 0.000 |
| 10 | 0.005 | 0.002 | 0.049 | 0.020 | 0.776 | 0.090 | 0.792 | 0.106 | 0.779 | 0.074 | 0.811 | 0.063 | 0.889 | 0.053 | 0.855 | 0.067 | 210.000 | 0.000 |
| 11 | 0.005 | 0.002 | 0.047 | 0.016 | 0.786 | 0.089 | 0.794 | 0.110 | 0.785 | 0.076 | 0.816 | 0.065 | 0.904 | 0.051 | 0.885 | 0.056 | 210.000 | 0.000 |
| 12 | 0.004 | 0.001 | 0.045 | 0.015 | 0.792 | 0.087 | 0.803 | 0.109 | 0.792 | 0.076 | 0.823 | 0.065 | 0.906 | 0.052 | 0.891 | 0.055 | 210.000 | 0.000 |
| 13 | 0.004 | 0.002 | 0.044 | 0.014 | 0.822 | 0.090 | 0.800 | 0.105 | 0.807 | 0.078 | 0.836 | 0.066 | 0.917 | 0.048 | 0.900 | 0.054 | 210.000 | 0.000 |
| 14 | 0.004 | 0.002 | 0.044 | 0.015 | 0.830 | 0.083 | 0.793 | 0.097 | 0.807 | 0.072 | 0.836 | 0.059 | 0.921 | 0.045 | 0.907 | 0.050 | 210.000 | 0.000 |
| 15 | 0.005 | 0.002 | 0.047 | 0.017 | 0.831 | 0.081 | 0.805 | 0.095 | 0.813 | 0.066 | 0.841 | 0.054 | 0.921 | 0.046 | 0.909 | 0.049 | 210.000 | 0.000 |
Using only new features:
0%| | 0/1 [00:00<?, ?it/s]
100%|██████████| 1/1 [00:00<00:00, 1004.86it/s]
0%| | 0/2 [00:00<?, ?it/s]
100%|██████████| 2/2 [00:00<00:00, 26.82it/s]
0%| | 0/3 [00:00<?, ?it/s]
100%|██████████| 3/3 [00:00<00:00, 18.15it/s]
100%|██████████| 3/3 [00:00<00:00, 17.56it/s]
0%| | 0/4 [00:00<?, ?it/s]
50%|█████ | 2/4 [00:00<00:00, 18.02it/s]
100%|██████████| 4/4 [00:00<00:00, 16.07it/s]
100%|██████████| 4/4 [00:00<00:00, 16.29it/s]
0%| | 0/5 [00:00<?, ?it/s]
60%|██████ | 3/5 [00:00<00:00, 25.63it/s]
100%|██████████| 5/5 [00:00<00:00, 21.90it/s]
0%| | 0/6 [00:00<?, ?it/s]
50%|█████ | 3/6 [00:00<00:00, 25.89it/s]
100%|██████████| 6/6 [00:00<00:00, 20.51it/s]
100%|██████████| 6/6 [00:00<00:00, 21.01it/s]
0%| | 0/7 [00:00<?, ?it/s]
43%|████▎ | 3/7 [00:00<00:00, 29.56it/s]
86%|████████▌ | 6/7 [00:00<00:00, 24.45it/s]
100%|██████████| 7/7 [00:00<00:00, 24.69it/s]
0%| | 0/8 [00:00<?, ?it/s]
25%|██▌ | 2/8 [00:00<00:00, 18.78it/s]
50%|█████ | 4/8 [00:00<00:00, 13.35it/s]
75%|███████▌ | 6/8 [00:00<00:00, 11.84it/s]
100%|██████████| 8/8 [00:00<00:00, 11.21it/s]
100%|██████████| 8/8 [00:00<00:00, 11.90it/s]
0%| | 0/9 [00:00<?, ?it/s]
22%|██▏ | 2/9 [00:00<00:00, 18.18it/s]
44%|████▍ | 4/9 [00:00<00:00, 11.32it/s]
67%|██████▋ | 6/9 [00:00<00:00, 9.66it/s]
89%|████████▉ | 8/9 [00:00<00:00, 9.45it/s]
100%|██████████| 9/9 [00:00<00:00, 9.93it/s]
0%| | 0/10 [00:00<?, ?it/s]
30%|███ | 3/10 [00:00<00:00, 16.85it/s]
50%|█████ | 5/10 [00:00<00:00, 14.48it/s]
70%|███████ | 7/10 [00:00<00:00, 13.91it/s]
90%|█████████ | 9/10 [00:00<00:00, 13.84it/s]
100%|██████████| 10/10 [00:00<00:00, 14.14it/s]
0%| | 0/11 [00:00<?, ?it/s]
27%|██▋ | 3/11 [00:00<00:00, 21.72it/s]
55%|█████▍ | 6/11 [00:00<00:00, 16.61it/s]
73%|███████▎ | 8/11 [00:00<00:00, 16.72it/s]
91%|█████████ | 10/11 [00:00<00:00, 16.73it/s]
100%|██████████| 11/11 [00:00<00:00, 17.28it/s]
0%| | 0/12 [00:00<?, ?it/s]
25%|██▌ | 3/12 [00:00<00:00, 24.26it/s]
50%|█████ | 6/12 [00:00<00:00, 18.78it/s]
67%|██████▋ | 8/12 [00:00<00:00, 18.24it/s]
83%|████████▎ | 10/12 [00:00<00:00, 17.54it/s]
100%|██████████| 12/12 [00:00<00:00, 18.05it/s]
100%|██████████| 12/12 [00:00<00:00, 18.39it/s]
0%| | 0/13 [00:00<?, ?it/s]
23%|██▎ | 3/13 [00:00<00:00, 23.39it/s]
46%|████▌ | 6/13 [00:00<00:00, 17.00it/s]
62%|██████▏ | 8/13 [00:00<00:00, 15.43it/s]
77%|███████▋ | 10/13 [00:00<00:00, 13.36it/s]
92%|█████████▏| 12/13 [00:00<00:00, 12.62it/s]
100%|██████████| 13/13 [00:00<00:00, 13.86it/s]
0%| | 0/14 [00:00<?, ?it/s]
21%|██▏ | 3/14 [00:00<00:00, 28.90it/s]
43%|████▎ | 6/14 [00:00<00:00, 21.43it/s]
64%|██████▍ | 9/14 [00:00<00:00, 21.57it/s]
86%|████████▌ | 12/14 [00:00<00:00, 21.69it/s]
100%|██████████| 14/14 [00:00<00:00, 21.91it/s]
0%| | 0/15 [00:00<?, ?it/s]
20%|██ | 3/15 [00:00<00:00, 21.03it/s]
40%|████ | 6/15 [00:00<00:00, 17.28it/s]
53%|█████▎ | 8/15 [00:00<00:00, 15.27it/s]
73%|███████▎ | 11/15 [00:00<00:00, 16.31it/s]
87%|████████▋ | 13/15 [00:00<00:00, 15.83it/s]
100%|██████████| 15/15 [00:00<00:00, 15.66it/s]
100%|██████████| 15/15 [00:00<00:00, 16.11it/s]
| fit_time | score_time | test_precision | test_recall | test_f1 | test_balanced_accuracy | test_roc_auc | test_average_precision | n_observations | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| mean | std | mean | std | mean | std | mean | std | mean | std | mean | std | mean | std | mean | std | mean | std | |
| n_features | ||||||||||||||||||
| 1 | 0.004 | 0.002 | 0.048 | 0.016 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.500 | 0.000 | 0.757 | 0.063 | 0.700 | 0.084 | 210.000 | 0.000 |
| 2 | 0.003 | 0.001 | 0.033 | 0.006 | 0.060 | 0.240 | 0.005 | 0.020 | 0.009 | 0.036 | 0.502 | 0.010 | 0.615 | 0.121 | 0.568 | 0.113 | 210.000 | 0.000 |
| 3 | 0.005 | 0.003 | 0.049 | 0.015 | 0.498 | 0.144 | 0.211 | 0.081 | 0.286 | 0.089 | 0.523 | 0.042 | 0.601 | 0.070 | 0.534 | 0.073 | 210.000 | 0.000 |
| 4 | 0.004 | 0.002 | 0.046 | 0.015 | 0.579 | 0.128 | 0.301 | 0.106 | 0.385 | 0.100 | 0.567 | 0.053 | 0.619 | 0.068 | 0.571 | 0.075 | 210.000 | 0.000 |
| 5 | 0.004 | 0.002 | 0.046 | 0.017 | 0.558 | 0.121 | 0.304 | 0.100 | 0.381 | 0.093 | 0.559 | 0.047 | 0.624 | 0.068 | 0.572 | 0.076 | 210.000 | 0.000 |
| 6 | 0.004 | 0.002 | 0.045 | 0.016 | 0.523 | 0.109 | 0.293 | 0.099 | 0.367 | 0.093 | 0.548 | 0.049 | 0.614 | 0.062 | 0.551 | 0.072 | 210.000 | 0.000 |
| 7 | 0.005 | 0.003 | 0.049 | 0.023 | 0.682 | 0.097 | 0.592 | 0.112 | 0.629 | 0.091 | 0.694 | 0.070 | 0.770 | 0.058 | 0.703 | 0.072 | 210.000 | 0.000 |
| 8 | 0.005 | 0.002 | 0.049 | 0.018 | 0.685 | 0.082 | 0.640 | 0.117 | 0.655 | 0.083 | 0.711 | 0.061 | 0.809 | 0.053 | 0.752 | 0.066 | 210.000 | 0.000 |
| 9 | 0.006 | 0.003 | 0.062 | 0.026 | 0.678 | 0.085 | 0.630 | 0.108 | 0.647 | 0.078 | 0.704 | 0.060 | 0.809 | 0.054 | 0.752 | 0.067 | 210.000 | 0.000 |
| 10 | 0.004 | 0.002 | 0.047 | 0.016 | 0.684 | 0.085 | 0.643 | 0.108 | 0.657 | 0.082 | 0.712 | 0.062 | 0.814 | 0.052 | 0.759 | 0.062 | 210.000 | 0.000 |
| 11 | 0.004 | 0.002 | 0.044 | 0.021 | 0.694 | 0.084 | 0.655 | 0.100 | 0.667 | 0.070 | 0.719 | 0.053 | 0.810 | 0.052 | 0.755 | 0.062 | 210.000 | 0.000 |
| 12 | 0.003 | 0.001 | 0.034 | 0.011 | 0.692 | 0.083 | 0.657 | 0.107 | 0.667 | 0.072 | 0.719 | 0.053 | 0.809 | 0.052 | 0.756 | 0.061 | 210.000 | 0.000 |
| 13 | 0.004 | 0.002 | 0.045 | 0.014 | 0.686 | 0.080 | 0.655 | 0.111 | 0.663 | 0.075 | 0.715 | 0.055 | 0.806 | 0.053 | 0.753 | 0.061 | 210.000 | 0.000 |
| 14 | 0.005 | 0.002 | 0.045 | 0.017 | 0.680 | 0.079 | 0.652 | 0.111 | 0.658 | 0.073 | 0.712 | 0.052 | 0.802 | 0.054 | 0.747 | 0.064 | 210.000 | 0.000 |
| 15 | 0.004 | 0.001 | 0.034 | 0.005 | 0.670 | 0.089 | 0.645 | 0.130 | 0.648 | 0.093 | 0.704 | 0.065 | 0.795 | 0.058 | 0.739 | 0.068 | 210.000 | 0.000 |
Best number of features by subset of the data:#
| ald | all | new | |
|---|---|---|---|
| fit_time | 8 | 7 | 9 |
| score_time | 8 | 7 | 9 |
| test_precision | 13 | 15 | 11 |
| test_recall | 12 | 9 | 12 |
| test_f1 | 13 | 15 | 11 |
| test_balanced_accuracy | 13 | 15 | 11 |
| test_roc_auc | 13 | 15 | 10 |
| test_average_precision | 13 | 15 | 10 |
| n_observations | 1 | 1 | 1 |
Train, test split#
Show number of cases in train and test data
| train | test | |
|---|---|---|
| False | 98 | 24 |
| True | 70 | 18 |
Results#
run_modelreturns dataclasses with the further needed resultsadd mrmr selection of data (select best number of features to use instead of fixing it)
Save results for final model on entire data, new features and ALD study criteria selected data.
0%| | 0/15 [00:00<?, ?it/s]
13%|█▎ | 2/15 [00:00<00:01, 6.71it/s]
20%|██ | 3/15 [00:00<00:02, 4.77it/s]
27%|██▋ | 4/15 [00:00<00:02, 4.12it/s]
33%|███▎ | 5/15 [00:01<00:02, 3.85it/s]
40%|████ | 6/15 [00:01<00:02, 3.75it/s]
47%|████▋ | 7/15 [00:01<00:02, 3.48it/s]
53%|█████▎ | 8/15 [00:02<00:01, 3.55it/s]
60%|██████ | 9/15 [00:02<00:01, 3.75it/s]
67%|██████▋ | 10/15 [00:02<00:01, 3.62it/s]
73%|███████▎ | 11/15 [00:02<00:01, 3.75it/s]
80%|████████ | 12/15 [00:03<00:00, 3.85it/s]
87%|████████▋ | 13/15 [00:03<00:00, 3.92it/s]
93%|█████████▎| 14/15 [00:03<00:00, 4.09it/s]
100%|██████████| 15/15 [00:03<00:00, 4.26it/s]
100%|██████████| 15/15 [00:03<00:00, 3.99it/s]
0%| | 0/10 [00:00<?, ?it/s]
40%|████ | 4/10 [00:00<00:00, 31.35it/s]
80%|████████ | 8/10 [00:00<00:00, 26.16it/s]
100%|██████████| 10/10 [00:00<00:00, 20.39it/s]
0%| | 0/13 [00:00<?, ?it/s]
15%|█▌ | 2/13 [00:00<00:01, 9.20it/s]
23%|██▎ | 3/13 [00:00<00:01, 6.58it/s]
31%|███ | 4/13 [00:00<00:01, 5.87it/s]
38%|███▊ | 5/13 [00:00<00:01, 5.33it/s]
46%|████▌ | 6/13 [00:01<00:01, 4.96it/s]
54%|█████▍ | 7/13 [00:01<00:01, 4.74it/s]
62%|██████▏ | 8/13 [00:01<00:00, 5.04it/s]
69%|██████▉ | 9/13 [00:01<00:00, 4.92it/s]
77%|███████▋ | 10/13 [00:01<00:00, 4.65it/s]
85%|████████▍ | 11/13 [00:02<00:00, 4.60it/s]
92%|█████████▏| 12/13 [00:02<00:00, 4.93it/s]
100%|██████████| 13/13 [00:02<00:00, 5.02it/s]
100%|██████████| 13/13 [00:02<00:00, 5.15it/s]
ROC-AUC on test split#
pimmslearn.plotting - INFO Saved Figures to runs/alzheimer_study/diff_analysis/AD/PI_vs_DAE/auc_roc_curve.pdf
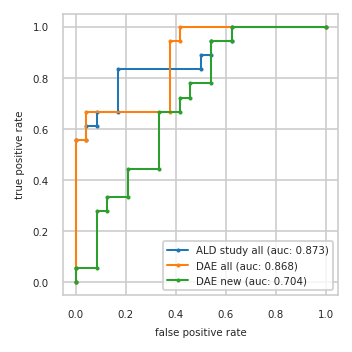
Data used to plot ROC:
| ALD study all | DAE all | DAE new | ||||
|---|---|---|---|---|---|---|
| fpr | tpr | fpr | tpr | fpr | tpr | |
| 0 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 |
| 1 | 0.000 | 0.056 | 0.000 | 0.056 | 0.000 | 0.056 |
| 2 | 0.000 | 0.556 | 0.000 | 0.556 | 0.083 | 0.056 |
| 3 | 0.042 | 0.556 | 0.042 | 0.556 | 0.083 | 0.278 |
| 4 | 0.042 | 0.611 | 0.042 | 0.667 | 0.125 | 0.278 |
| 5 | 0.083 | 0.611 | 0.375 | 0.667 | 0.125 | 0.333 |
| 6 | 0.083 | 0.667 | 0.375 | 0.944 | 0.208 | 0.333 |
| 7 | 0.167 | 0.667 | 0.417 | 0.944 | 0.208 | 0.444 |
| 8 | 0.167 | 0.833 | 0.417 | 1.000 | 0.333 | 0.444 |
| 9 | 0.500 | 0.833 | 1.000 | 1.000 | 0.333 | 0.667 |
| 10 | 0.500 | 0.889 | NaN | NaN | 0.417 | 0.667 |
| 11 | 0.542 | 0.889 | NaN | NaN | 0.417 | 0.722 |
| 12 | 0.542 | 0.944 | NaN | NaN | 0.458 | 0.722 |
| 13 | 0.625 | 0.944 | NaN | NaN | 0.458 | 0.778 |
| 14 | 0.625 | 1.000 | NaN | NaN | 0.542 | 0.778 |
| 15 | 1.000 | 1.000 | NaN | NaN | 0.542 | 0.944 |
| 16 | NaN | NaN | NaN | NaN | 0.625 | 0.944 |
| 17 | NaN | NaN | NaN | NaN | 0.625 | 1.000 |
| 18 | NaN | NaN | NaN | NaN | 1.000 | 1.000 |
Features selected for final models#
| ALD study all | DAE all | DAE new | |
|---|---|---|---|
| rank | |||
| 0 | P10636-2;P10636-6 | P10636-2;P10636-6 | Q14894 |
| 1 | Q6MZW2 | Q8NBI6 | P01704 |
| 2 | P02741 | P31946;P31946-2 | P51688 |
| 3 | Q13231;Q13231-3 | Q9Y2T3;Q9Y2T3-3 | P31321 |
| 4 | P61981 | P61981 | A0A0C4DGV4;E9PLX3;O43504;R4GMU8 |
| 5 | P04075 | C9JF17;P05090 | Q96GD0 |
| 6 | P14174 | P04075 | F8WBF9;Q5TH30;Q9UGV2;Q9UGV2-2;Q9UGV2-3 |
| 7 | P14618 | P63104 | E5RJY1;E7ESM1;Q92597;Q92597-2;Q92597-3 |
| 8 | P08294 | P15151-2 | Q9NUQ9 |
| 9 | P00338;P00338-3 | Q14894 | A0A1W2PQ94;B4DS77;B4DS77-2;B4DS77-3 |
| 10 | P05160 | P14174 | None |
| 11 | Q6EMK4 | A0A0C4DGY8;D6RA00;Q9UHY7 | None |
| 12 | C9JF17;P05090 | P00338;P00338-3 | None |
| 13 | None | P00492 | None |
| 14 | None | Q6EMK4 | None |
Precision-Recall plot on test data#
pimmslearn.plotting - INFO Saved Figures to runs/alzheimer_study/diff_analysis/AD/PI_vs_DAE/prec_recall_curve.pdf
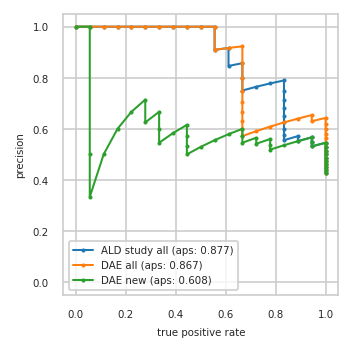
Data used to plot PRC:
| ALD study all | DAE all | DAE new | ||||
|---|---|---|---|---|---|---|
| precision | tpr | precision | tpr | precision | tpr | |
| 0 | 0.429 | 1.000 | 0.429 | 1.000 | 0.429 | 1.000 |
| 1 | 0.439 | 1.000 | 0.439 | 1.000 | 0.439 | 1.000 |
| 2 | 0.450 | 1.000 | 0.450 | 1.000 | 0.450 | 1.000 |
| 3 | 0.462 | 1.000 | 0.462 | 1.000 | 0.462 | 1.000 |
| 4 | 0.474 | 1.000 | 0.474 | 1.000 | 0.474 | 1.000 |
| 5 | 0.486 | 1.000 | 0.486 | 1.000 | 0.486 | 1.000 |
| 6 | 0.500 | 1.000 | 0.500 | 1.000 | 0.500 | 1.000 |
| 7 | 0.514 | 1.000 | 0.514 | 1.000 | 0.514 | 1.000 |
| 8 | 0.529 | 1.000 | 0.529 | 1.000 | 0.529 | 1.000 |
| 9 | 0.545 | 1.000 | 0.545 | 1.000 | 0.545 | 1.000 |
| 10 | 0.531 | 0.944 | 0.562 | 1.000 | 0.531 | 0.944 |
| 11 | 0.548 | 0.944 | 0.581 | 1.000 | 0.548 | 0.944 |
| 12 | 0.567 | 0.944 | 0.600 | 1.000 | 0.567 | 0.944 |
| 13 | 0.552 | 0.889 | 0.621 | 1.000 | 0.552 | 0.889 |
| 14 | 0.571 | 0.889 | 0.643 | 1.000 | 0.536 | 0.833 |
| 15 | 0.556 | 0.833 | 0.630 | 0.944 | 0.519 | 0.778 |
| 16 | 0.577 | 0.833 | 0.654 | 0.944 | 0.538 | 0.778 |
| 17 | 0.600 | 0.833 | 0.640 | 0.889 | 0.560 | 0.778 |
| 18 | 0.625 | 0.833 | 0.625 | 0.833 | 0.542 | 0.722 |
| 19 | 0.652 | 0.833 | 0.609 | 0.778 | 0.565 | 0.722 |
| 20 | 0.682 | 0.833 | 0.591 | 0.722 | 0.545 | 0.667 |
| 21 | 0.714 | 0.833 | 0.571 | 0.667 | 0.571 | 0.667 |
| 22 | 0.750 | 0.833 | 0.600 | 0.667 | 0.600 | 0.667 |
| 23 | 0.789 | 0.833 | 0.632 | 0.667 | 0.579 | 0.611 |
| 24 | 0.778 | 0.778 | 0.667 | 0.667 | 0.556 | 0.556 |
| 25 | 0.765 | 0.722 | 0.706 | 0.667 | 0.529 | 0.500 |
| 26 | 0.750 | 0.667 | 0.750 | 0.667 | 0.500 | 0.444 |
| 27 | 0.800 | 0.667 | 0.800 | 0.667 | 0.533 | 0.444 |
| 28 | 0.857 | 0.667 | 0.857 | 0.667 | 0.571 | 0.444 |
| 29 | 0.846 | 0.611 | 0.923 | 0.667 | 0.615 | 0.444 |
| 30 | 0.917 | 0.611 | 0.917 | 0.611 | 0.583 | 0.389 |
| 31 | 0.909 | 0.556 | 0.909 | 0.556 | 0.545 | 0.333 |
| 32 | 1.000 | 0.556 | 1.000 | 0.556 | 0.600 | 0.333 |
| 33 | 1.000 | 0.500 | 1.000 | 0.500 | 0.667 | 0.333 |
| 34 | 1.000 | 0.444 | 1.000 | 0.444 | 0.625 | 0.278 |
| 35 | 1.000 | 0.389 | 1.000 | 0.389 | 0.714 | 0.278 |
| 36 | 1.000 | 0.333 | 1.000 | 0.333 | 0.667 | 0.222 |
| 37 | 1.000 | 0.278 | 1.000 | 0.278 | 0.600 | 0.167 |
| 38 | 1.000 | 0.222 | 1.000 | 0.222 | 0.500 | 0.111 |
| 39 | 1.000 | 0.167 | 1.000 | 0.167 | 0.333 | 0.056 |
| 40 | 1.000 | 0.111 | 1.000 | 0.111 | 0.500 | 0.056 |
| 41 | 1.000 | 0.056 | 1.000 | 0.056 | 1.000 | 0.056 |
| 42 | 1.000 | 0.000 | 1.000 | 0.000 | 1.000 | 0.000 |
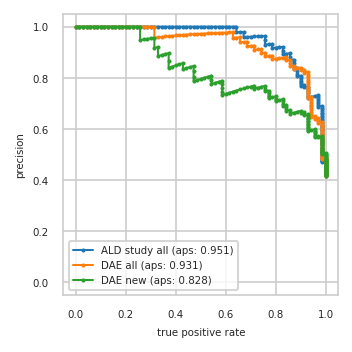
Train data plots#
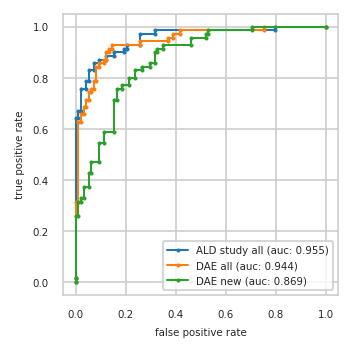
pimmslearn.plotting - INFO Saved Figures to runs/alzheimer_study/diff_analysis/AD/PI_vs_DAE/prec_recall_curve_train.pdf
pimmslearn.plotting - INFO Saved Figures to runs/alzheimer_study/diff_analysis/AD/PI_vs_DAE/auc_roc_curve_train.pdf
Output files:
{'results_DAE all.pkl': PosixPath('runs/alzheimer_study/diff_analysis/AD/PI_vs_DAE/results_DAE all.pkl'),
'results_DAE new.pkl': PosixPath('runs/alzheimer_study/diff_analysis/AD/PI_vs_DAE/results_DAE new.pkl'),
'results_ALD study all.pkl': PosixPath('runs/alzheimer_study/diff_analysis/AD/PI_vs_DAE/results_ALD study all.pkl'),
'auc_roc_curve.pdf': PosixPath('runs/alzheimer_study/diff_analysis/AD/PI_vs_DAE/auc_roc_curve.pdf'),
'mrmr_feat_by_model.xlsx': PosixPath('runs/alzheimer_study/diff_analysis/AD/PI_vs_DAE/mrmr_feat_by_model.xlsx'),
'prec_recall_curve.pdf': PosixPath('runs/alzheimer_study/diff_analysis/AD/PI_vs_DAE/prec_recall_curve.pdf'),
'prec_recall_curve_train.pdf': PosixPath('runs/alzheimer_study/diff_analysis/AD/PI_vs_DAE/prec_recall_curve_train.pdf'),
'auc_roc_curve_train.pdf': PosixPath('runs/alzheimer_study/diff_analysis/AD/PI_vs_DAE/auc_roc_curve_train.pdf')}