Compare outcomes from differential analysis based on different imputation methods#
load scores based on
10_1_ald_diff_analysis
Parameters#
Default and set parameters for the notebook.
folder_experiment = 'runs/appl_ald_data/plasma/proteinGroups'
target = 'kleiner'
model_key = 'VAE'
baseline = 'RSN'
out_folder = 'diff_analysis'
selected_statistics = ['p-unc', '-Log10 pvalue', 'qvalue', 'rejected']
disease_ontology = 5082 # code from https://disease-ontology.org/
# split diseases notebook? Query gene names for proteins in file from uniprot?
annotaitons_gene_col = 'PG.Genes'
# Parameters
disease_ontology = 10652
folder_experiment = "runs/alzheimer_study"
target = "AD"
baseline = "PI"
model_key = "QRILC"
out_folder = "diff_analysis"
annotaitons_gene_col = "None"
Add set parameters to configuration
root - INFO Removed from global namespace: folder_experiment
root - INFO Removed from global namespace: target
root - INFO Removed from global namespace: model_key
root - INFO Removed from global namespace: baseline
root - INFO Removed from global namespace: out_folder
root - INFO Removed from global namespace: selected_statistics
root - INFO Removed from global namespace: disease_ontology
root - INFO Removed from global namespace: annotaitons_gene_col
root - INFO Already set attribute: folder_experiment has value runs/alzheimer_study
root - INFO Already set attribute: out_folder has value diff_analysis
{'annotaitons_gene_col': 'None',
'baseline': 'PI',
'data': PosixPath('runs/alzheimer_study/data'),
'disease_ontology': 10652,
'folder_experiment': PosixPath('runs/alzheimer_study'),
'freq_features_observed': PosixPath('runs/alzheimer_study/freq_features_observed.csv'),
'model_key': 'QRILC',
'out_figures': PosixPath('runs/alzheimer_study/figures'),
'out_folder': PosixPath('runs/alzheimer_study/diff_analysis/AD/PI_vs_QRILC'),
'out_metrics': PosixPath('runs/alzheimer_study'),
'out_models': PosixPath('runs/alzheimer_study'),
'out_preds': PosixPath('runs/alzheimer_study/preds'),
'scores_folder': PosixPath('runs/alzheimer_study/diff_analysis/AD/scores'),
'selected_statistics': ['p-unc', '-Log10 pvalue', 'qvalue', 'rejected'],
'target': 'AD'}
Excel file for exports#
files_out = dict()
writer_args = dict(float_format='%.3f')
fname = args.out_folder / 'diff_analysis_compare_methods.xlsx'
files_out[fname.name] = fname
writer = pd.ExcelWriter(fname)
logger.info("Writing to excel file: %s", fname)
root - INFO Writing to excel file: runs/alzheimer_study/diff_analysis/AD/PI_vs_QRILC/diff_analysis_compare_methods.xlsx
Load scores#
Load baseline model scores#
Show all statistics, later use selected statistics
| model | PI | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| var | SS | DF | F | p-unc | np2 | -Log10 pvalue | qvalue | rejected | |
| protein groups | Source | ||||||||
| A0A024QZX5;A0A087X1N8;P35237 | AD | 0.180 | 1 | 0.291 | 0.590 | 0.002 | 0.229 | 0.728 | False |
| age | 0.084 | 1 | 0.135 | 0.713 | 0.001 | 0.147 | 0.817 | False | |
| Kiel | 2.216 | 1 | 3.588 | 0.060 | 0.018 | 1.224 | 0.140 | False | |
| Magdeburg | 5.950 | 1 | 9.634 | 0.002 | 0.048 | 2.657 | 0.010 | True | |
| Sweden | 10.249 | 1 | 16.595 | 0.000 | 0.080 | 4.169 | 0.000 | True | |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| S4R3U6 | AD | 0.367 | 1 | 0.390 | 0.533 | 0.002 | 0.273 | 0.680 | False |
| age | 1.453 | 1 | 1.542 | 0.216 | 0.008 | 0.666 | 0.365 | False | |
| Kiel | 0.099 | 1 | 0.105 | 0.746 | 0.001 | 0.127 | 0.840 | False | |
| Magdeburg | 3.254 | 1 | 3.453 | 0.065 | 0.018 | 1.189 | 0.148 | False | |
| Sweden | 8.790 | 1 | 9.329 | 0.003 | 0.047 | 2.589 | 0.011 | True | |
7105 rows × 8 columns
Load selected comparison model scores#
| model | QRILC | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| var | SS | DF | F | p-unc | np2 | -Log10 pvalue | qvalue | rejected | |
| protein groups | Source | ||||||||
| A0A024QZX5;A0A087X1N8;P35237 | AD | 0.836 | 1 | 4.919 | 0.028 | 0.025 | 1.557 | 0.071 | False |
| age | 0.008 | 1 | 0.048 | 0.826 | 0.000 | 0.083 | 0.891 | False | |
| Kiel | 0.444 | 1 | 2.612 | 0.108 | 0.013 | 0.968 | 0.209 | False | |
| Magdeburg | 0.981 | 1 | 5.771 | 0.017 | 0.029 | 1.763 | 0.048 | True | |
| Sweden | 2.458 | 1 | 14.455 | 0.000 | 0.070 | 3.714 | 0.001 | True | |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| S4R3U6 | AD | 0.881 | 1 | 0.462 | 0.498 | 0.002 | 0.303 | 0.637 | False |
| age | 0.714 | 1 | 0.374 | 0.541 | 0.002 | 0.266 | 0.672 | False | |
| Kiel | 7.750 | 1 | 4.059 | 0.045 | 0.021 | 1.344 | 0.105 | False | |
| Magdeburg | 19.996 | 1 | 10.474 | 0.001 | 0.052 | 2.846 | 0.006 | True | |
| Sweden | 0.678 | 1 | 0.355 | 0.552 | 0.002 | 0.258 | 0.681 | False | |
7105 rows × 8 columns
Combined scores#
show only selected statistics for comparsion
| model | PI | QRILC | |||||||
|---|---|---|---|---|---|---|---|---|---|
| var | p-unc | -Log10 pvalue | qvalue | rejected | p-unc | -Log10 pvalue | qvalue | rejected | |
| protein groups | Source | ||||||||
| A0A024QZX5;A0A087X1N8;P35237 | AD | 0.590 | 0.229 | 0.728 | False | 0.028 | 1.557 | 0.071 | False |
| Kiel | 0.060 | 1.224 | 0.140 | False | 0.108 | 0.968 | 0.209 | False | |
| Magdeburg | 0.002 | 2.657 | 0.010 | True | 0.017 | 1.763 | 0.048 | True | |
| Sweden | 0.000 | 4.169 | 0.000 | True | 0.000 | 3.714 | 0.001 | True | |
| age | 0.713 | 0.147 | 0.817 | False | 0.826 | 0.083 | 0.891 | False | |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| S4R3U6 | AD | 0.533 | 0.273 | 0.680 | False | 0.498 | 0.303 | 0.637 | False |
| Kiel | 0.746 | 0.127 | 0.840 | False | 0.045 | 1.344 | 0.105 | False | |
| Magdeburg | 0.065 | 1.189 | 0.148 | False | 0.001 | 2.846 | 0.006 | True | |
| Sweden | 0.003 | 2.589 | 0.011 | True | 0.552 | 0.258 | 0.681 | False | |
| age | 0.216 | 0.666 | 0.365 | False | 0.541 | 0.266 | 0.672 | False | |
7105 rows × 8 columns
Models in comparison (name mapping)
{'PI': 'PI', 'QRILC': 'QRILC'}
Describe scores#
| model | PI | QRILC | ||||
|---|---|---|---|---|---|---|
| var | p-unc | -Log10 pvalue | qvalue | p-unc | -Log10 pvalue | qvalue |
| count | 7,105.000 | 7,105.000 | 7,105.000 | 7,105.000 | 7,105.000 | 7,105.000 |
| mean | 0.261 | 2.486 | 0.338 | 0.246 | 2.735 | 0.313 |
| std | 0.304 | 5.378 | 0.332 | 0.299 | 5.167 | 0.327 |
| min | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 |
| 25% | 0.004 | 0.327 | 0.015 | 0.002 | 0.358 | 0.008 |
| 50% | 0.119 | 0.925 | 0.238 | 0.093 | 1.031 | 0.186 |
| 75% | 0.471 | 2.440 | 0.628 | 0.439 | 2.698 | 0.585 |
| max | 1.000 | 143.804 | 1.000 | 0.999 | 83.512 | 0.999 |
One to one comparison of by feature:#
/tmp/ipykernel_102245/3761369923.py:2: FutureWarning: Starting with pandas version 3.0 all arguments of to_excel except for the argument 'excel_writer' will be keyword-only.
scores.to_excel(writer, 'scores', **writer_args)
| model | PI | QRILC | |||||||
|---|---|---|---|---|---|---|---|---|---|
| var | p-unc | -Log10 pvalue | qvalue | rejected | p-unc | -Log10 pvalue | qvalue | rejected | |
| protein groups | Source | ||||||||
| A0A024QZX5;A0A087X1N8;P35237 | AD | 0.590 | 0.229 | 0.728 | False | 0.028 | 1.557 | 0.071 | False |
| A0A024R0T9;K7ER74;P02655 | AD | 0.059 | 1.228 | 0.139 | False | 0.030 | 1.517 | 0.076 | False |
| A0A024R3W6;A0A024R412;O60462;O60462-2;O60462-3;O60462-4;O60462-5;Q7LBX6;X5D2Q8 | AD | 0.114 | 0.944 | 0.230 | False | 0.190 | 0.721 | 0.321 | False |
| A0A024R644;A0A0A0MRU5;A0A1B0GWI2;O75503 | AD | 0.524 | 0.281 | 0.672 | False | 0.314 | 0.503 | 0.466 | False |
| A0A075B6H7 | AD | 0.109 | 0.964 | 0.223 | False | 0.112 | 0.952 | 0.214 | False |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| Q9Y6R7 | AD | 0.175 | 0.756 | 0.315 | False | 0.175 | 0.756 | 0.302 | False |
| Q9Y6X5 | AD | 0.050 | 1.305 | 0.121 | False | 0.076 | 1.120 | 0.159 | False |
| Q9Y6Y8;Q9Y6Y8-2 | AD | 0.083 | 1.079 | 0.181 | False | 0.083 | 1.079 | 0.171 | False |
| Q9Y6Y9 | AD | 0.302 | 0.520 | 0.464 | False | 0.495 | 0.305 | 0.635 | False |
| S4R3U6 | AD | 0.533 | 0.273 | 0.680 | False | 0.498 | 0.303 | 0.637 | False |
1421 rows × 8 columns
And the descriptive statistics of the numeric values:
| model | PI | QRILC | ||||
|---|---|---|---|---|---|---|
| var | p-unc | -Log10 pvalue | qvalue | p-unc | -Log10 pvalue | qvalue |
| count | 1,421.000 | 1,421.000 | 1,421.000 | 1,421.000 | 1,421.000 | 1,421.000 |
| mean | 0.254 | 1.405 | 0.336 | 0.251 | 1.486 | 0.324 |
| std | 0.292 | 1.623 | 0.319 | 0.291 | 1.765 | 0.317 |
| min | 0.000 | 0.001 | 0.000 | 0.000 | 0.002 | 0.000 |
| 25% | 0.011 | 0.352 | 0.036 | 0.009 | 0.353 | 0.030 |
| 50% | 0.125 | 0.904 | 0.247 | 0.109 | 0.961 | 0.211 |
| 75% | 0.444 | 1.960 | 0.605 | 0.443 | 2.023 | 0.589 |
| max | 0.997 | 23.616 | 0.998 | 0.995 | 23.394 | 0.996 |
and the boolean decision values
| model | PI | QRILC |
|---|---|---|
| var | rejected | rejected |
| count | 1421 | 1421 |
| unique | 2 | 2 |
| top | False | False |
| freq | 1027 | 996 |
Load frequencies of observed features#
| data | |
|---|---|
| frequency | |
| protein groups | |
| A0A024QZX5;A0A087X1N8;P35237 | 186 |
| A0A024R0T9;K7ER74;P02655 | 195 |
| A0A024R3W6;A0A024R412;O60462;O60462-2;O60462-3;O60462-4;O60462-5;Q7LBX6;X5D2Q8 | 174 |
| A0A024R644;A0A0A0MRU5;A0A1B0GWI2;O75503 | 196 |
| A0A075B6H7 | 91 |
| ... | ... |
| Q9Y6R7 | 197 |
| Q9Y6X5 | 173 |
| Q9Y6Y8;Q9Y6Y8-2 | 197 |
| Q9Y6Y9 | 119 |
| S4R3U6 | 126 |
1421 rows × 1 columns
Plot qvalues of both models with annotated decisions#
Prepare data for plotting (qvalues)
| PI | QRILC | frequency | Differential Analysis Comparison | |
|---|---|---|---|---|
| protein groups | ||||
| A0A024QZX5;A0A087X1N8;P35237 | 0.728 | 0.071 | 186 | PI (no) - QRILC (no) |
| A0A024R0T9;K7ER74;P02655 | 0.139 | 0.076 | 195 | PI (no) - QRILC (no) |
| A0A024R3W6;A0A024R412;O60462;O60462-2;O60462-3;O60462-4;O60462-5;Q7LBX6;X5D2Q8 | 0.230 | 0.321 | 174 | PI (no) - QRILC (no) |
| A0A024R644;A0A0A0MRU5;A0A1B0GWI2;O75503 | 0.672 | 0.466 | 196 | PI (no) - QRILC (no) |
| A0A075B6H7 | 0.223 | 0.214 | 91 | PI (no) - QRILC (no) |
| ... | ... | ... | ... | ... |
| Q9Y6R7 | 0.315 | 0.302 | 197 | PI (no) - QRILC (no) |
| Q9Y6X5 | 0.121 | 0.159 | 173 | PI (no) - QRILC (no) |
| Q9Y6Y8;Q9Y6Y8-2 | 0.181 | 0.171 | 197 | PI (no) - QRILC (no) |
| Q9Y6Y9 | 0.464 | 0.635 | 119 | PI (no) - QRILC (no) |
| S4R3U6 | 0.680 | 0.637 | 126 | PI (no) - QRILC (no) |
1421 rows × 4 columns
List of features with the highest difference in qvalues
| PI | QRILC | frequency | Differential Analysis Comparison | diff_qvalue | |
|---|---|---|---|---|---|
| protein groups | |||||
| E7EN89;E9PP67;E9PQ25;F2Z2Y8;Q9H0E2;Q9H0E2-2 | 0.940 | 0.003 | 86 | PI (no) - QRILC (yes) | 0.936 |
| Q8TEA8 | 0.016 | 0.917 | 56 | PI (yes) - QRILC (no) | 0.901 |
| P43004;P43004-2;P43004-3 | 0.839 | 0.016 | 89 | PI (no) - QRILC (yes) | 0.823 |
| P35754 | 0.036 | 0.807 | 143 | PI (yes) - QRILC (no) | 0.772 |
| A0A087WTT8;A0A0A0MQX5;O94779;O94779-2 | 0.004 | 0.504 | 114 | PI (yes) - QRILC (no) | 0.500 |
| ... | ... | ... | ... | ... | ... |
| P26572 | 0.056 | 0.048 | 194 | PI (no) - QRILC (yes) | 0.007 |
| D6RCE0;E9PD25;O43897;O43897-2 | 0.050 | 0.044 | 180 | PI (no) - QRILC (yes) | 0.006 |
| Q16706 | 0.052 | 0.046 | 195 | PI (no) - QRILC (yes) | 0.006 |
| P00740;P00740-2 | 0.053 | 0.048 | 197 | PI (no) - QRILC (yes) | 0.004 |
| K7ERG9;P00746 | 0.052 | 0.048 | 197 | PI (no) - QRILC (yes) | 0.004 |
109 rows × 5 columns
Differences plotted with created annotations#
pimmslearn.plotting - INFO Saved Figures to runs/alzheimer_study/diff_analysis/AD/PI_vs_QRILC/diff_analysis_comparision_1_QRILC
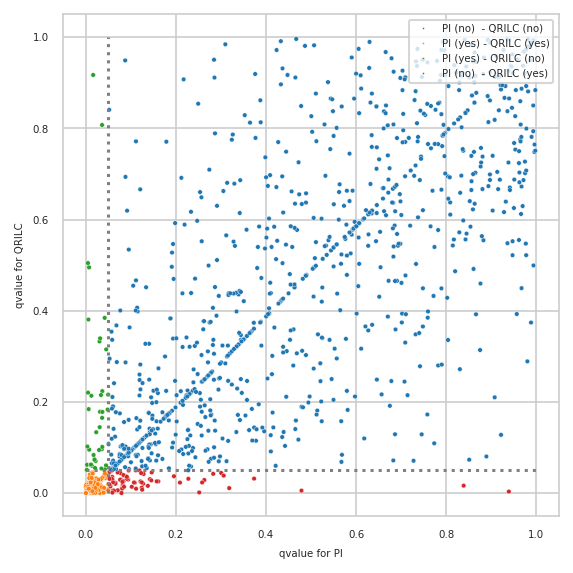
also showing how many features were measured (“observed”) by size of circle
pimmslearn.plotting - INFO Saved Figures to runs/alzheimer_study/diff_analysis/AD/PI_vs_QRILC/diff_analysis_comparision_2_QRILC

Only features contained in model#
this block exist due to a specific part in the ALD analysis of the paper
root - INFO No features only in new comparision model.
DISEASES DB lookup#
Query diseases database for gene associations with specified disease ontology id.
pimmslearn.databases.diseases - WARNING There are more associations available
| ENSP | score | |
|---|---|---|
| None | ||
| APOE | ENSP00000252486 | 5.000 |
| PSEN1 | ENSP00000326366 | 5.000 |
| PSEN2 | ENSP00000355747 | 5.000 |
| APP | ENSP00000284981 | 5.000 |
| TREM2 | ENSP00000362205 | 4.825 |
| ... | ... | ... |
| CARMIL1 | ENSP00000331983 | 0.681 |
| CENPJ | ENSP00000371308 | 0.681 |
| ERP27 | ENSP00000266397 | 0.681 |
| ZNF585B | ENSP00000433773 | 0.681 |
| KIR3DL2 | ENSP00000325525 | 0.681 |
10000 rows × 2 columns
only by model#
Only by model which were significant#
Only significant by RSN#
mask = (scores_common[(str(args.baseline), 'rejected')] & mask_different)
mask.sum()