Transfer predictions from NAGuideR#
Papermill script parameters:
# files and folders
# Datasplit folder with data for experiment
folder_experiment: str = 'runs/example'
folder_data: str = '' # specify data directory if needed
file_format: str = 'csv' # file format of create splits, default pickle (csv)
identifer_str: str = '_all_' # identifier for prediction files to be filtered
dumps: list = None # list of dumps to be used
# Parameters
dumps = "runs/alzheimer_study/preds/pred_all_BPCA.csv,runs/alzheimer_study/preds/pred_all_COLMEDIAN.csv,runs/alzheimer_study/preds/pred_all_IMPSEQ.csv,runs/alzheimer_study/preds/pred_all_IMPSEQROB.csv,runs/alzheimer_study/preds/pred_all_IRM.csv,runs/alzheimer_study/preds/pred_all_KNN_IMPUTE.csv,runs/alzheimer_study/preds/pred_all_LLS.csv,runs/alzheimer_study/preds/pred_all_MINDET.csv,runs/alzheimer_study/preds/pred_all_MINIMUM.csv,runs/alzheimer_study/preds/pred_all_MINPROB.csv,runs/alzheimer_study/preds/pred_all_MLE.csv,runs/alzheimer_study/preds/pred_all_PI.csv,runs/alzheimer_study/preds/pred_all_QRILC.csv,runs/alzheimer_study/preds/pred_all_RF.csv,runs/alzheimer_study/preds/pred_all_ROWMEDIAN.csv,runs/alzheimer_study/preds/pred_all_SVDMETHOD.csv,runs/alzheimer_study/preds/pred_all_TRKNN.csv,runs/alzheimer_study/preds/pred_all_ZERO.csv"
folder_experiment = "runs/alzheimer_study"
Some argument transformations
{'data': Path('runs/alzheimer_study/data'),
'dumps': 'runs/alzheimer_study/preds/pred_all_BPCA.csv,runs/alzheimer_study/preds/pred_all_COLMEDIAN.csv,runs/alzheimer_study/preds/pred_all_IMPSEQ.csv,runs/alzheimer_study/preds/pred_all_IMPSEQROB.csv,runs/alzheimer_study/preds/pred_all_IRM.csv,runs/alzheimer_study/preds/pred_all_KNN_IMPUTE.csv,runs/alzheimer_study/preds/pred_all_LLS.csv,runs/alzheimer_study/preds/pred_all_MINDET.csv,runs/alzheimer_study/preds/pred_all_MINIMUM.csv,runs/alzheimer_study/preds/pred_all_MINPROB.csv,runs/alzheimer_study/preds/pred_all_MLE.csv,runs/alzheimer_study/preds/pred_all_PI.csv,runs/alzheimer_study/preds/pred_all_QRILC.csv,runs/alzheimer_study/preds/pred_all_RF.csv,runs/alzheimer_study/preds/pred_all_ROWMEDIAN.csv,runs/alzheimer_study/preds/pred_all_SVDMETHOD.csv,runs/alzheimer_study/preds/pred_all_TRKNN.csv,runs/alzheimer_study/preds/pred_all_ZERO.csv',
'file_format': 'csv',
'folder_data': '',
'folder_experiment': Path('runs/alzheimer_study'),
'identifer_str': '_all_',
'out_figures': Path('runs/alzheimer_study/figures'),
'out_folder': Path('runs/alzheimer_study'),
'out_metrics': Path('runs/alzheimer_study'),
'out_models': Path('runs/alzheimer_study'),
'out_preds': Path('runs/alzheimer_study/preds')}
load data splits
pimmslearn.io.datasplits - INFO Loaded 'train_X' from file: runs/alzheimer_study/data/train_X.csv
pimmslearn.io.datasplits - INFO Loaded 'val_y' from file: runs/alzheimer_study/data/val_y.csv
pimmslearn.io.datasplits - INFO Loaded 'test_y' from file: runs/alzheimer_study/data/test_y.csv
Validation and test data split of simulated missing values
| observed | ||
|---|---|---|
| Sample ID | protein groups | |
| Sample_158 | Q9UN70;Q9UN70-2 | 14.630 |
| Sample_050 | Q9Y287 | 15.755 |
| Sample_107 | Q8N475;Q8N475-2 | 15.029 |
| Sample_199 | P06307 | 19.376 |
| Sample_067 | Q5VUB5 | 15.309 |
| ... | ... | ... |
| Sample_111 | F6SYF8;Q9UBP4 | 22.822 |
| Sample_002 | A0A0A0MT36 | 18.165 |
| Sample_049 | Q8WY21;Q8WY21-2;Q8WY21-3;Q8WY21-4 | 15.525 |
| Sample_182 | Q8NFT8 | 14.379 |
| Sample_123 | Q16853;Q16853-2 | 14.504 |
12600 rows × 1 columns
| observed | |
|---|---|
| count | 12,600.000 |
| mean | 16.339 |
| std | 2.741 |
| min | 7.209 |
| 25% | 14.412 |
| 50% | 15.935 |
| 75% | 17.910 |
| max | 30.140 |
[Path('runs/alzheimer_study/preds/pred_all_BPCA.csv'),
Path('runs/alzheimer_study/preds/pred_all_COLMEDIAN.csv'),
Path('runs/alzheimer_study/preds/pred_all_IMPSEQ.csv'),
Path('runs/alzheimer_study/preds/pred_all_IMPSEQROB.csv'),
Path('runs/alzheimer_study/preds/pred_all_IRM.csv'),
Path('runs/alzheimer_study/preds/pred_all_KNN_IMPUTE.csv'),
Path('runs/alzheimer_study/preds/pred_all_LLS.csv'),
Path('runs/alzheimer_study/preds/pred_all_MINDET.csv'),
Path('runs/alzheimer_study/preds/pred_all_MINIMUM.csv'),
Path('runs/alzheimer_study/preds/pred_all_MINPROB.csv'),
Path('runs/alzheimer_study/preds/pred_all_MLE.csv'),
Path('runs/alzheimer_study/preds/pred_all_PI.csv'),
Path('runs/alzheimer_study/preds/pred_all_QRILC.csv'),
Path('runs/alzheimer_study/preds/pred_all_RF.csv'),
Path('runs/alzheimer_study/preds/pred_all_ROWMEDIAN.csv'),
Path('runs/alzheimer_study/preds/pred_all_SVDMETHOD.csv'),
Path('runs/alzheimer_study/preds/pred_all_TRKNN.csv'),
Path('runs/alzheimer_study/preds/pred_all_ZERO.csv')]
pimmslearn - INFO Load fpath = Path('runs/alzheimer_study/preds/pred_all_BPCA.csv')
pimmslearn - INFO Save fname = Path('runs/alzheimer_study/preds/pred_val_BPCA.csv')
pimmslearn - INFO Save fname = Path('runs/alzheimer_study/preds/pred_test_BPCA.csv')
pimmslearn - INFO Save fname = Path('runs/alzheimer_study/preds/pred_real_na_BPCA.csv')
pimmslearn - INFO Load fpath = Path('runs/alzheimer_study/preds/pred_all_COLMEDIAN.csv')
pimmslearn - INFO Save fname = Path('runs/alzheimer_study/preds/pred_val_COLMEDIAN.csv')
pimmslearn - INFO Save fname = Path('runs/alzheimer_study/preds/pred_test_COLMEDIAN.csv')
pimmslearn - INFO Save fname = Path('runs/alzheimer_study/preds/pred_real_na_COLMEDIAN.csv')
pimmslearn - INFO Load fpath = Path('runs/alzheimer_study/preds/pred_all_IMPSEQ.csv')
pimmslearn - INFO Save fname = Path('runs/alzheimer_study/preds/pred_val_IMPSEQ.csv')
pimmslearn - INFO Save fname = Path('runs/alzheimer_study/preds/pred_test_IMPSEQ.csv')
pimmslearn - INFO Save fname = Path('runs/alzheimer_study/preds/pred_real_na_IMPSEQ.csv')
pimmslearn - INFO Load fpath = Path('runs/alzheimer_study/preds/pred_all_IMPSEQROB.csv')
pimmslearn - INFO Save fname = Path('runs/alzheimer_study/preds/pred_val_IMPSEQROB.csv')
pimmslearn - INFO Save fname = Path('runs/alzheimer_study/preds/pred_test_IMPSEQROB.csv')
pimmslearn - INFO Save fname = Path('runs/alzheimer_study/preds/pred_real_na_IMPSEQROB.csv')
pimmslearn - INFO Load fpath = Path('runs/alzheimer_study/preds/pred_all_IRM.csv')
pimmslearn - INFO Save fname = Path('runs/alzheimer_study/preds/pred_val_IRM.csv')
pimmslearn - INFO Save fname = Path('runs/alzheimer_study/preds/pred_test_IRM.csv')
pimmslearn - INFO Save fname = Path('runs/alzheimer_study/preds/pred_real_na_IRM.csv')
pimmslearn - INFO Load fpath = Path('runs/alzheimer_study/preds/pred_all_KNN_IMPUTE.csv')
pimmslearn - INFO Save fname = Path('runs/alzheimer_study/preds/pred_val_KNN_IMPUTE.csv')
pimmslearn - INFO Save fname = Path('runs/alzheimer_study/preds/pred_test_KNN_IMPUTE.csv')
pimmslearn - INFO Save fname = Path('runs/alzheimer_study/preds/pred_real_na_KNN_IMPUTE.csv')
pimmslearn - INFO Load fpath = Path('runs/alzheimer_study/preds/pred_all_LLS.csv')
pimmslearn - INFO Save fname = Path('runs/alzheimer_study/preds/pred_val_LLS.csv')
pimmslearn - INFO Save fname = Path('runs/alzheimer_study/preds/pred_test_LLS.csv')
pimmslearn - INFO Save fname = Path('runs/alzheimer_study/preds/pred_real_na_LLS.csv')
pimmslearn - INFO Load fpath = Path('runs/alzheimer_study/preds/pred_all_MINDET.csv')
pimmslearn - INFO Save fname = Path('runs/alzheimer_study/preds/pred_val_MINDET.csv')
pimmslearn - INFO Save fname = Path('runs/alzheimer_study/preds/pred_test_MINDET.csv')
pimmslearn - INFO Save fname = Path('runs/alzheimer_study/preds/pred_real_na_MINDET.csv')
pimmslearn - INFO Load fpath = Path('runs/alzheimer_study/preds/pred_all_MINIMUM.csv')
pimmslearn - INFO Save fname = Path('runs/alzheimer_study/preds/pred_val_MINIMUM.csv')
pimmslearn - INFO Save fname = Path('runs/alzheimer_study/preds/pred_test_MINIMUM.csv')
pimmslearn - INFO Save fname = Path('runs/alzheimer_study/preds/pred_real_na_MINIMUM.csv')
pimmslearn - INFO Load fpath = Path('runs/alzheimer_study/preds/pred_all_MINPROB.csv')
pimmslearn - INFO Save fname = Path('runs/alzheimer_study/preds/pred_val_MINPROB.csv')
pimmslearn - INFO Save fname = Path('runs/alzheimer_study/preds/pred_test_MINPROB.csv')
pimmslearn - INFO Save fname = Path('runs/alzheimer_study/preds/pred_real_na_MINPROB.csv')
pimmslearn - INFO Load fpath = Path('runs/alzheimer_study/preds/pred_all_MLE.csv')
pimmslearn - INFO Save fname = Path('runs/alzheimer_study/preds/pred_val_MLE.csv')
pimmslearn - INFO Save fname = Path('runs/alzheimer_study/preds/pred_test_MLE.csv')
pimmslearn - INFO Save fname = Path('runs/alzheimer_study/preds/pred_real_na_MLE.csv')
pimmslearn - INFO Load fpath = Path('runs/alzheimer_study/preds/pred_all_PI.csv')
pimmslearn - INFO Save fname = Path('runs/alzheimer_study/preds/pred_val_PI.csv')
pimmslearn - INFO Save fname = Path('runs/alzheimer_study/preds/pred_test_PI.csv')
pimmslearn - INFO Save fname = Path('runs/alzheimer_study/preds/pred_real_na_PI.csv')
pimmslearn - INFO Load fpath = Path('runs/alzheimer_study/preds/pred_all_QRILC.csv')
pimmslearn - INFO Save fname = Path('runs/alzheimer_study/preds/pred_val_QRILC.csv')
pimmslearn - INFO Save fname = Path('runs/alzheimer_study/preds/pred_test_QRILC.csv')
pimmslearn - INFO Save fname = Path('runs/alzheimer_study/preds/pred_real_na_QRILC.csv')
pimmslearn - INFO Load fpath = Path('runs/alzheimer_study/preds/pred_all_RF.csv')
pimmslearn - INFO Save fname = Path('runs/alzheimer_study/preds/pred_val_RF.csv')
pimmslearn - INFO Save fname = Path('runs/alzheimer_study/preds/pred_test_RF.csv')
pimmslearn - INFO Save fname = Path('runs/alzheimer_study/preds/pred_real_na_RF.csv')
pimmslearn - INFO Load fpath = Path('runs/alzheimer_study/preds/pred_all_ROWMEDIAN.csv')
pimmslearn - INFO Save fname = Path('runs/alzheimer_study/preds/pred_val_ROWMEDIAN.csv')
pimmslearn - INFO Save fname = Path('runs/alzheimer_study/preds/pred_test_ROWMEDIAN.csv')
pimmslearn - INFO Save fname = Path('runs/alzheimer_study/preds/pred_real_na_ROWMEDIAN.csv')
pimmslearn - INFO Load fpath = Path('runs/alzheimer_study/preds/pred_all_SVDMETHOD.csv')
pimmslearn - INFO Save fname = Path('runs/alzheimer_study/preds/pred_val_SVDMETHOD.csv')
pimmslearn - INFO Save fname = Path('runs/alzheimer_study/preds/pred_test_SVDMETHOD.csv')
pimmslearn - INFO Save fname = Path('runs/alzheimer_study/preds/pred_real_na_SVDMETHOD.csv')
pimmslearn - INFO Load fpath = Path('runs/alzheimer_study/preds/pred_all_TRKNN.csv')
pimmslearn - INFO Save fname = Path('runs/alzheimer_study/preds/pred_val_TRKNN.csv')
pimmslearn - INFO Save fname = Path('runs/alzheimer_study/preds/pred_test_TRKNN.csv')
pimmslearn - INFO Save fname = Path('runs/alzheimer_study/preds/pred_real_na_TRKNN.csv')
pimmslearn - INFO Load fpath = Path('runs/alzheimer_study/preds/pred_all_ZERO.csv')
pimmslearn - INFO Save fname = Path('runs/alzheimer_study/preds/pred_val_ZERO.csv')
pimmslearn - INFO Save fname = Path('runs/alzheimer_study/preds/pred_test_ZERO.csv')
pimmslearn - INFO Save fname = Path('runs/alzheimer_study/preds/pred_real_na_ZERO.csv')
| observed | BPCA | COLMEDIAN | IMPSEQ | IMPSEQROB | IRM | KNN_IMPUTE | LLS | MINDET | MINIMUM | MINPROB | MLE | PI | QRILC | RF | ROWMEDIAN | SVDMETHOD | TRKNN | ZERO | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sample ID | protein groups | |||||||||||||||||||
| Sample_158 | Q9UN70;Q9UN70-2 | 14.630 | 15.469 | 16.800 | NaN | 58.276 | 15.871 | 15.937 | 15.858 | 11.916 | 7.068 | 10.929 | 2,513.638 | 11.373 | 14.631 | 15.597 | 15.752 | 17.206 | 15.700 | 0 |
| Sample_050 | Q9Y287 | 15.755 | 16.453 | 17.288 | NaN | 16.993 | 17.472 | 16.961 | 17.058 | 12.900 | 7.068 | 12.593 | 19.829 | 12.117 | 16.151 | 16.846 | 17.221 | 17.807 | 16.738 | 0 |
| Sample_107 | Q8N475;Q8N475-2 | 15.029 | 13.110 | 17.187 | NaN | -78.084 | 12.680 | 15.437 | 14.397 | 12.313 | 7.068 | 11.907 | 2,582.130 | 12.354 | 13.316 | 14.311 | 14.846 | 17.434 | 13.776 | 0 |
| Sample_199 | P06307 | 19.376 | 19.639 | 16.711 | NaN | 102.283 | 19.632 | 18.861 | 19.005 | 12.285 | 7.068 | 12.106 | 2,483.120 | 12.446 | 17.005 | 18.988 | 18.973 | 17.111 | 19.015 | 0 |
| Sample_067 | Q5VUB5 | 15.309 | 15.465 | 16.743 | NaN | -36.470 | 15.976 | 15.079 | 15.104 | 11.827 | 7.068 | 12.167 | 2,569.564 | 13.566 | 12.305 | 15.002 | 14.726 | 17.031 | 14.699 | 0 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| Sample_111 | F6SYF8;Q9UBP4 | 22.822 | 22.994 | 17.042 | NaN | 104.484 | 22.983 | 22.837 | 22.836 | 12.161 | 7.068 | 12.722 | 2,634.108 | 14.010 | 22.019 | 22.851 | 22.918 | 17.330 | 22.872 | 0 |
| Sample_002 | A0A0A0MT36 | 18.165 | 15.882 | 16.792 | NaN | -18.408 | 17.237 | 15.446 | 15.579 | 12.586 | 7.068 | 12.925 | 2,448.503 | 12.662 | 13.329 | 15.823 | 15.877 | 16.879 | 15.671 | 0 |
| Sample_049 | Q8WY21;Q8WY21-2;Q8WY21-3;Q8WY21-4 | 15.525 | 15.406 | 17.032 | NaN | -27.128 | 15.313 | 15.995 | 15.903 | 12.352 | 7.068 | 12.760 | 2,487.550 | 13.417 | 14.969 | 15.851 | 16.278 | 17.215 | 15.574 | 0 |
| Sample_182 | Q8NFT8 | 14.379 | 14.322 | 16.764 | NaN | -12.434 | 14.873 | 14.675 | 13.456 | 12.504 | 7.068 | 13.018 | 2,426.191 | 11.854 | 12.500 | 13.996 | 13.995 | 17.125 | 14.518 | 0 |
| Sample_123 | Q16853;Q16853-2 | 14.504 | 14.582 | 16.686 | NaN | 78.799 | 14.748 | 14.824 | 14.931 | 12.689 | 7.068 | 13.310 | 2,461.806 | 13.023 | 13.923 | 14.757 | 14.849 | 16.981 | 14.485 | 0 |
12600 rows × 19 columns
Metrics for simulated missing values (NA)
Selected as truth to compare to: observed
| BPCA | COLMEDIAN | IMPSEQROB | IRM | KNN_IMPUTE | LLS | MINDET | MINIMUM | MINPROB | MLE | PI | QRILC | RF | ROWMEDIAN | SVDMETHOD | TRKNN | ZERO | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MSE | 0.429 | 7.554 | 741,117.141 | 0.751 | 0.753 | 3,023.655 | 23.897 | 93.346 | 24.346 | 5,469,016.990 | 21.471 | 4.374 | 0.494 | 0.766 | 8.005 | 0.469 | 274.368 |
| MAE | 0.422 | 2.210 | 333.478 | 0.588 | 0.554 | 1.329 | 4.108 | 9.272 | 4.136 | 2,172.384 | 3.808 | 1.651 | 0.461 | 0.598 | 2.309 | 0.450 | 16.340 |
| N | 12,600.000 | 12,600.000 | 12,600.000 | 12,600.000 | 12,600.000 | 12,600.000 | 12,600.000 | 12,600.000 | 12,600.000 | 12,600.000 | 12,600.000 | 12,600.000 | 12,600.000 | 12,600.000 | 12,600.000 | 12,600.000 | 12,600.000 |
| prop | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 |
Test Datasplit#
Selected as truth to compare to: observed
| BPCA | COLMEDIAN | IMPSEQROB | IRM | KNN_IMPUTE | LLS | MINDET | MINIMUM | MINPROB | MLE | PI | QRILC | RF | ROWMEDIAN | SVDMETHOD | TRKNN | ZERO | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MSE | 0.455 | 7.704 | 741,503.932 | 0.763 | 0.772 | 838.462 | 23.999 | 93.458 | 24.381 | 5,509,003.848 | 21.619 | 4.264 | 0.515 | 0.776 | 8.159 | 0.500 | 274.464 |
| MAE | 0.432 | 2.223 | 334.546 | 0.587 | 0.558 | 0.874 | 4.109 | 9.271 | 4.127 | 2,186.302 | 3.804 | 1.630 | 0.465 | 0.602 | 2.330 | 0.458 | 16.339 |
| N | 12,600.000 | 12,600.000 | 12,600.000 | 12,600.000 | 12,600.000 | 12,600.000 | 12,600.000 | 12,600.000 | 12,600.000 | 12,600.000 | 12,600.000 | 12,600.000 | 12,600.000 | 12,600.000 | 12,600.000 | 12,600.000 | 12,600.000 |
| prop | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 |
| subset | valid_fake_na | test_fake_na | |
|---|---|---|---|
| model | metric_name | ||
| BPCA | MSE | 0.429 | 0.455 |
| MAE | 0.422 | 0.432 | |
| N | 12,600.000 | 12,600.000 | |
| prop | 1.000 | 1.000 | |
| COLMEDIAN | MSE | 7.554 | 7.704 |
| ... | ... | ... | ... |
| TRKNN | prop | 1.000 | 1.000 |
| ZERO | MSE | 274.368 | 274.464 |
| MAE | 16.340 | 16.339 | |
| N | 12,600.000 | 12,600.000 | |
| prop | 1.000 | 1.000 |
68 rows × 2 columns
model metric_name
BPCA MAE 0.422
TRKNN MAE 0.450
RF MAE 0.461
KNN_IMPUTE MAE 0.554
IRM MAE 0.588
ROWMEDIAN MAE 0.598
LLS MAE 1.329
QRILC MAE 1.651
COLMEDIAN MAE 2.210
SVDMETHOD MAE 2.309
PI MAE 3.808
MINDET MAE 4.108
MINPROB MAE 4.136
MINIMUM MAE 9.272
ZERO MAE 16.340
IMPSEQROB MAE 333.478
MLE MAE 2,172.384
Name: valid_fake_na, dtype: float64
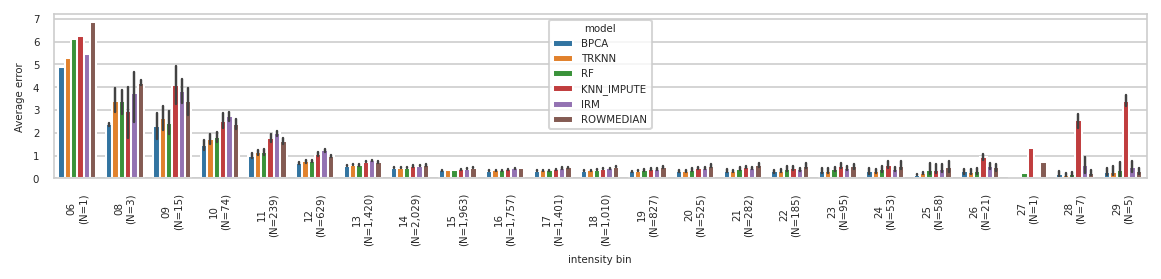
['observed', 'BPCA', 'TRKNN', 'RF', 'KNN_IMPUTE', 'IRM', 'ROWMEDIAN']
pimmslearn.plotting - INFO Saved Figures to runs/alzheimer_study/figures/NAGuideR_errors_per_bin_val.png
/home/runner/work/pimms/pimms/project/.snakemake/conda/924ec7e362d761ecf0807b9074d79999_/lib/python3.12/site-packages/pimmslearn/plotting/errors.py:45: FutureWarning:
The `errwidth` parameter is deprecated. And will be removed in v0.15.0. Pass `err_kws={'linewidth': 1.2}` instead.
ax = sns.barplot(data=errors_binned, ax=ax,
{'pred_val_BPCA.csv': 'runs/alzheimer_study/preds/pred_val_BPCA.csv',
'pred_test_BPCA.csv': 'runs/alzheimer_study/preds/pred_test_BPCA.csv',
'pred_real_na_BPCA.csv': 'runs/alzheimer_study/preds/pred_real_na_BPCA.csv',
'pred_val_COLMEDIAN.csv': 'runs/alzheimer_study/preds/pred_val_COLMEDIAN.csv',
'pred_test_COLMEDIAN.csv': 'runs/alzheimer_study/preds/pred_test_COLMEDIAN.csv',
'pred_real_na_COLMEDIAN.csv': 'runs/alzheimer_study/preds/pred_real_na_COLMEDIAN.csv',
'pred_val_IMPSEQ.csv': 'runs/alzheimer_study/preds/pred_val_IMPSEQ.csv',
'pred_test_IMPSEQ.csv': 'runs/alzheimer_study/preds/pred_test_IMPSEQ.csv',
'pred_real_na_IMPSEQ.csv': 'runs/alzheimer_study/preds/pred_real_na_IMPSEQ.csv',
'pred_val_IMPSEQROB.csv': 'runs/alzheimer_study/preds/pred_val_IMPSEQROB.csv',
'pred_test_IMPSEQROB.csv': 'runs/alzheimer_study/preds/pred_test_IMPSEQROB.csv',
'pred_real_na_IMPSEQROB.csv': 'runs/alzheimer_study/preds/pred_real_na_IMPSEQROB.csv',
'pred_val_IRM.csv': 'runs/alzheimer_study/preds/pred_val_IRM.csv',
'pred_test_IRM.csv': 'runs/alzheimer_study/preds/pred_test_IRM.csv',
'pred_real_na_IRM.csv': 'runs/alzheimer_study/preds/pred_real_na_IRM.csv',
'pred_val_KNN_IMPUTE.csv': 'runs/alzheimer_study/preds/pred_val_KNN_IMPUTE.csv',
'pred_test_KNN_IMPUTE.csv': 'runs/alzheimer_study/preds/pred_test_KNN_IMPUTE.csv',
'pred_real_na_KNN_IMPUTE.csv': 'runs/alzheimer_study/preds/pred_real_na_KNN_IMPUTE.csv',
'pred_val_LLS.csv': 'runs/alzheimer_study/preds/pred_val_LLS.csv',
'pred_test_LLS.csv': 'runs/alzheimer_study/preds/pred_test_LLS.csv',
'pred_real_na_LLS.csv': 'runs/alzheimer_study/preds/pred_real_na_LLS.csv',
'pred_val_MINDET.csv': 'runs/alzheimer_study/preds/pred_val_MINDET.csv',
'pred_test_MINDET.csv': 'runs/alzheimer_study/preds/pred_test_MINDET.csv',
'pred_real_na_MINDET.csv': 'runs/alzheimer_study/preds/pred_real_na_MINDET.csv',
'pred_val_MINIMUM.csv': 'runs/alzheimer_study/preds/pred_val_MINIMUM.csv',
'pred_test_MINIMUM.csv': 'runs/alzheimer_study/preds/pred_test_MINIMUM.csv',
'pred_real_na_MINIMUM.csv': 'runs/alzheimer_study/preds/pred_real_na_MINIMUM.csv',
'pred_val_MINPROB.csv': 'runs/alzheimer_study/preds/pred_val_MINPROB.csv',
'pred_test_MINPROB.csv': 'runs/alzheimer_study/preds/pred_test_MINPROB.csv',
'pred_real_na_MINPROB.csv': 'runs/alzheimer_study/preds/pred_real_na_MINPROB.csv',
'pred_val_MLE.csv': 'runs/alzheimer_study/preds/pred_val_MLE.csv',
'pred_test_MLE.csv': 'runs/alzheimer_study/preds/pred_test_MLE.csv',
'pred_real_na_MLE.csv': 'runs/alzheimer_study/preds/pred_real_na_MLE.csv',
'pred_val_PI.csv': 'runs/alzheimer_study/preds/pred_val_PI.csv',
'pred_test_PI.csv': 'runs/alzheimer_study/preds/pred_test_PI.csv',
'pred_real_na_PI.csv': 'runs/alzheimer_study/preds/pred_real_na_PI.csv',
'pred_val_QRILC.csv': 'runs/alzheimer_study/preds/pred_val_QRILC.csv',
'pred_test_QRILC.csv': 'runs/alzheimer_study/preds/pred_test_QRILC.csv',
'pred_real_na_QRILC.csv': 'runs/alzheimer_study/preds/pred_real_na_QRILC.csv',
'pred_val_RF.csv': 'runs/alzheimer_study/preds/pred_val_RF.csv',
'pred_test_RF.csv': 'runs/alzheimer_study/preds/pred_test_RF.csv',
'pred_real_na_RF.csv': 'runs/alzheimer_study/preds/pred_real_na_RF.csv',
'pred_val_ROWMEDIAN.csv': 'runs/alzheimer_study/preds/pred_val_ROWMEDIAN.csv',
'pred_test_ROWMEDIAN.csv': 'runs/alzheimer_study/preds/pred_test_ROWMEDIAN.csv',
'pred_real_na_ROWMEDIAN.csv': 'runs/alzheimer_study/preds/pred_real_na_ROWMEDIAN.csv',
'pred_val_SVDMETHOD.csv': 'runs/alzheimer_study/preds/pred_val_SVDMETHOD.csv',
'pred_test_SVDMETHOD.csv': 'runs/alzheimer_study/preds/pred_test_SVDMETHOD.csv',
'pred_real_na_SVDMETHOD.csv': 'runs/alzheimer_study/preds/pred_real_na_SVDMETHOD.csv',
'pred_val_TRKNN.csv': 'runs/alzheimer_study/preds/pred_val_TRKNN.csv',
'pred_test_TRKNN.csv': 'runs/alzheimer_study/preds/pred_test_TRKNN.csv',
'pred_real_na_TRKNN.csv': 'runs/alzheimer_study/preds/pred_real_na_TRKNN.csv',
'pred_val_ZERO.csv': 'runs/alzheimer_study/preds/pred_val_ZERO.csv',
'pred_test_ZERO.csv': 'runs/alzheimer_study/preds/pred_test_ZERO.csv',
'pred_real_na_ZERO.csv': 'runs/alzheimer_study/preds/pred_real_na_ZERO.csv',
'NAGuideR_errors_per_bin_val.png': 'runs/alzheimer_study/figures/NAGuideR_errors_per_bin_val.png'}